Metabolic Fate of Modified Nucleotides after RNA Turnover, an Overlooked Issue in RNA Modification
Chen et al. reveal a multilayer molecular protection system functioning in Arabidopsis and human cells https://doi.org/10.1105/tpc.18.00236
By Mingjia Chen and Claus-Peter Witte
Background: RNA possesses over 100 distinct posttranscriptional modifications in eukaryotic species. N6-methyladenosine (m6A) is the most abundant internal modification in messenger RNAs. It modulates mRNA characteristics influencing RNA processing, translation, and RNA decay. The m6A patterns in mRNA are dynamically controlled by “writer” and “eraser” proteins in the nucleus, which act on specific sites of the mRNA. Quantitatively little methylation or demethylation on m6A mRNA occurs in the cytosol. Therefore, upon mRNA turnover in the cytosol, N6-methyl-AMP (N6-mAMP) will be released together with other mononucleotides.
Question: What is the metabolic fate of N6-mAMP resulting from RNA turnover? How does the cell prevent N6-mAMP from being recycled, as this may lead to misincorporation of m6A into newly forming RNA?
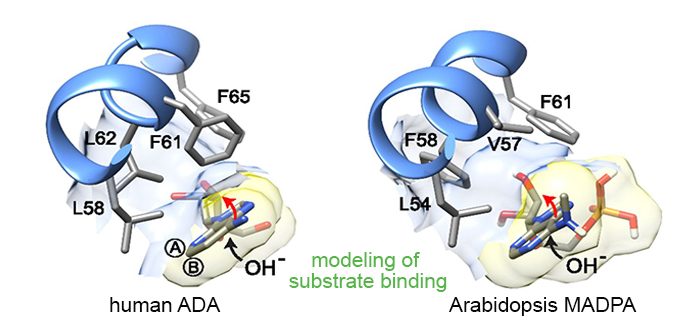
Findings: We identified an evolutionary conserved N6-mAMP deaminase (MAPDA), which hydrolyzes free N6-mAMP to inosine monophosphate in Arabidopsis and human cells. Upon abrogation of MAPDA, N6-methylated adenine derivatives accumulate in vivo, and plant root growth is slightly reduced. Together with adenylate kinases, which selectively phosphorylate AMP and have only low activity with N6-mAMP, MAPDA represents a molecular safeguard preventing the formation of N6-mATP. This is required because eukaryotic RNA polymerase II accepts N6-mATP as a substrate to a certain extent. Therefore, the accumulation of this compound may lead to random nonspecific incorporation of m6A marks into new RNA. However, in MAPDA mutants this effect was too small to be detected directly in the RNA, and it is conceivable that MAPDA has been conserved in most eukaryotes also because it protects other cellular processes from N6-methylated adenylates.
Next steps: Mutation of MAPDA also leads to N6-methyl adenosine accumulation. Which phosphatase catalyzes this reaction and are adenosine kinases selective against methylated adenosine? How are other modified nucleosides processed after RNA degradation? Do they accumulate or are they catabolized? Which enzymes are involved and what happens if they are abrogated?
Mingjia Chen, Mounashree J. Urs, Ismael Sánchez-González, Monilola A. Olayioye, Marco Herde, Claus-Peter Witte. (2018). m6A RNA degradation products are catabolized by an evolutionary conserved N6-methyl-AMP deaminase in plants and mammalian cells. Plant Cell https://doi.org/10.1105/tpc.18.00236.
Keywords: N6-mAMP, m6A, RNA epigenetics, adenylate kinase