CONSTANS Companion: CO Binds the NF-YB/NF-YC Dimer and Confers Sequence-Specific DNA Binding
One of the central players in the complex regulation of flowering, CONSTANS (CO) functions as a center for integration of the various signals that determine the timing of flowering. As output, CO regulates the expression of FLOWERING LOCUS T (FT) and other genes (reviewed by Shim et al., 2017). CO contains a CCT (CONSTANS, CONSTANS-LIKE, TOC1) domain similar to the conserved domain in NF-YA, a subunit of the Nuclear Factor-Y (NF-Y) transcription factor complex. NF-Y binds to a CCAAT box in the FT promoter and CO binds to a CO-Responsive Element (CORE) in the FT promoter.
NF-Y contains three subunits; two of these subunits, NF-YB and NF-YC, contain histone fold domains and interact through these domains to form a dimer, analogous to the dimer formed by histones H2B and H2A. The third subunit, NF-YA, binds this dimer and provides sequence specificity for binding to the CCAAT box. Animal genomes generally encode only one or two versions of each subunit and plant genomes have about a dozen genes for each subunit. Many of these genes have diverged in expression pattern, but show strong conservation of the histone fold domain, and a stretch in NF-YA required for sequence-specific binding of DNA and binding the histone fold domain.
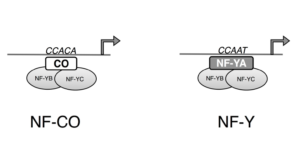 Work in Arabidopsis thaliana showed that NF-YB and NF-YC physically interact with CO. To examine whether CO forms a trimeric complex analogous to NF-Y, Gnesutta et al. (2017) conducted electrophoretic mobility shift assays (EMSAs) with the purified CO CCT domain, Arabidopsis NF-YB2 and NF-YC3, and a probe with the FT CORE2 sequence. Indeed the EMSAs showed that CO plus NF-YB2/NF-YC3 bound the CORE2 specifically, and did not bind the CCAAT box, and that CO alone showed little or no binding to CORE2. The authors named the CO/NF-YB2/NF-YC3 complex the NF-CO complex.
Work in Arabidopsis thaliana showed that NF-YB and NF-YC physically interact with CO. To examine whether CO forms a trimeric complex analogous to NF-Y, Gnesutta et al. (2017) conducted electrophoretic mobility shift assays (EMSAs) with the purified CO CCT domain, Arabidopsis NF-YB2 and NF-YC3, and a probe with the FT CORE2 sequence. Indeed the EMSAs showed that CO plus NF-YB2/NF-YC3 bound the CORE2 specifically, and did not bind the CCAAT box, and that CO alone showed little or no binding to CORE2. The authors named the CO/NF-YB2/NF-YC3 complex the NF-CO complex.
The authors next examined the preferred binding sequences of NF-CO. To this end, they examined the ability of mutant probes to compete for binding to the NF-CO complex by EMSA, and found that this complex prefers to bind the CORE element CCACA, with specific flanking sequences. The authors used RNA-sequencing analysis of co-sail, nf-yb, and nf-yc mutants to identify the genes potentially regulated by NF-CO, which included FT (as expected) and circadian regulation genes such as GIGANTEA; this analysis also refined the definition of the matrix of sequences NF-CO can potentially bind. Intriguingly, some genes that showed alterations in nf-yb and nf-yc mutants did not show alterations in the co-sail mutants, indicating that other proteins with CCT domains might interact with these NF-CO subunits. The Arabidopsis thaliana genome encodes forty proteins with predicted CCT domains, indicating many potential binding partners. Co-immunoprecipitation assays in the various mutants also showed that CO did not bind to the FT promoter in the absence of NF-YB2 and NF-YB3.
In the NF-Y complex, NF-YA interacts with a region formed by specific a helices of NF-YB and NF-YC. Analysis of NF-YB2 and NF-YC3 mutations in amino acids that are important for heterotrimer formation in NF-Y showed that these mutations affected formation of NF-CO, indicating that CO and NF-YA may bind to NF-YB/NF-YC in a similar way. Likewise, mutation of a conserved amino acid in CO abolished binding to CORE2. The similarities of binding indicate that CO and NF-YA might compete for binding, providing a possible explanation for the observation that overexpression of some NF-YAs can affect flowering, and yet another cautionary tale for interpretation of overexpression assays.
The large number of different NF-Y subunits, binding sites, and CCT domain-containing proteins presents a staggering number of potential complexes. Untangling their combinatorial activities and the multitude of processes where these complexes act, remain exciting possibilities for future research.
IN BRIEF by Jennifer Mach [email protected]
REFERENCES
Gnesutta, N., Kumimoto, R.W., Swain, S., Chiara, M., Siriwardana, C., Horner, D.S., Holt III, B.F., and Mantovani, R. (2017) CONSTANS imparts DNA sequence-specificity to the histone-fold NF-YB/NF-YC dimer. Plant Cell 10.1105/tpc.16.00864.
Shim, J.S., Kubota, A., and Imaizumi, T. (2017). Circadian Clock and Photoperiodic Flowering in Arabidopsis: CONSTANS Is a Hub for Signal Integration. Plant Physiol. 173, 5-15.



Leave a Reply
Want to join the discussion?Feel free to contribute!