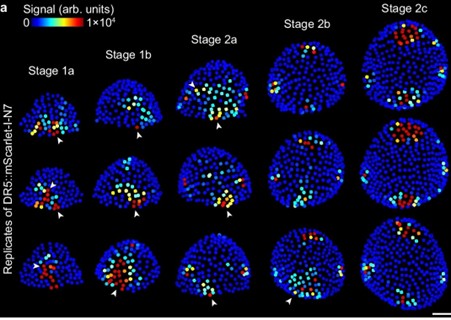
Stochastic gene expression, an ordinary part of multicellular life
Plant Science Research WeeklyStochasticity is an important feature of genes, allowing for variability in their abundance and activity prior to ‘fine-tuning’ at later periods of development. Kong et al. describe this feature in auxin responsive genes, attributing their ‘stochastic’- or inherently random expression – as…
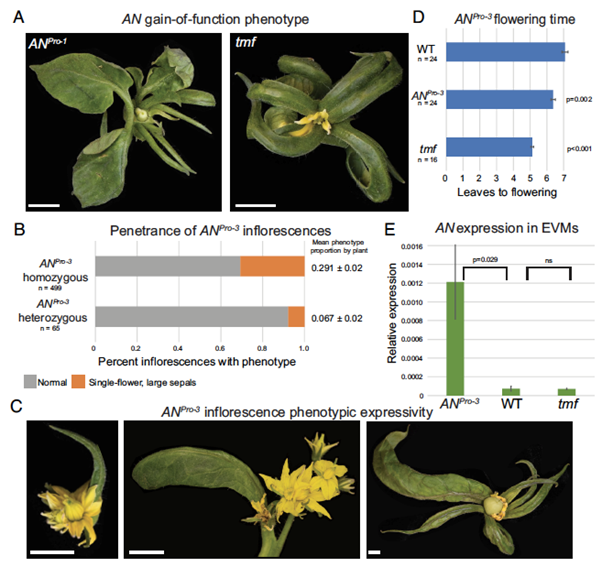
Developmental robustness from antagonizing cis-elements
Plant Science Research WeeklyDevelopmental transitions in plants are tightly regulated by transcriptional networks that require fine-tuned temporal and spatial control, with noncoding sequences in gene promoters playing a key role. The conserved transcriptional regulator UNUSUAL FLORAL ORGANS (UFO) is essential for floral development.…
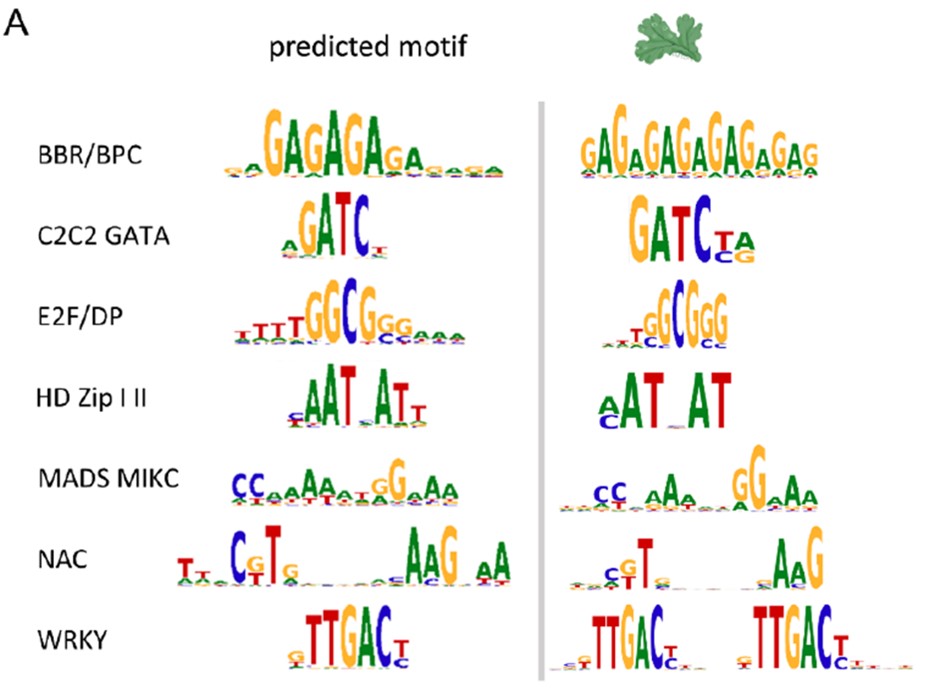
Many plant transcription factor families have evolutionarily conserved binding motifs
Plant Science Research WeeklyThe regulated expression of genes is fundamental to all biological processes, including development, cell growth, and responses to environmental signals. Transcription factors (TFs) are sequence-specific DNA-binding proteins that play a central role in transcriptional regulation by directly interacting…
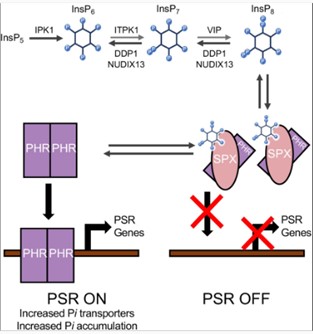
Disrupting plant inositol pyrophosphates to mitigate P pollution
Plant Science Research WeeklyInositol pyrophosphates (PP-InsP) are plant signaling molecules that regulate phosphate homeostasis and related metabolic processes. But a global phosphate crisis, due to resource depletion and environmental pollution from excess phosphate, has raised the need for phosphate management. Freed et al. investigated…
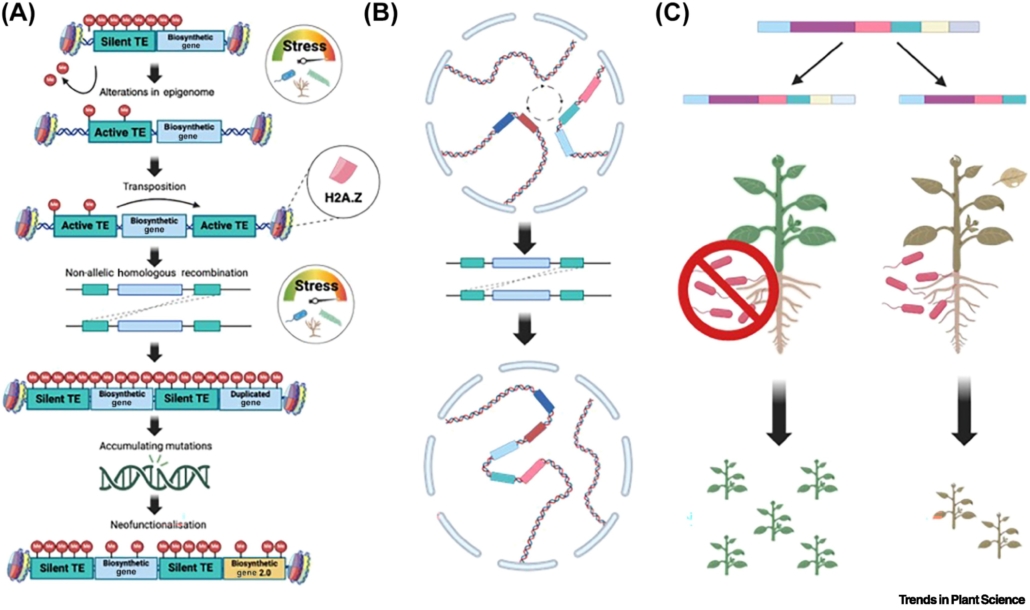
Review. Decoding resilience: Ecology, regulation, and evolution of biosynthetic gene clusters
Plant Science Research WeeklyAlthough clusters of functionally related genes are common in prokaryotes, until recently it was thought that they were not a feature of eukaryotic genomes. However, several studies have identified biosynthetic gene clusters (BGCs) in plants. Many of these gene clusters include sets of enzymes that act…

Mechanism of auxin-dependent gene regulation through composite auxin response elements
Plant Science Research WeeklyAuxin signaling influences plant growth and development by controlling gene expression, often through binding of transcription factors from the Auxin Response Factor (ARF) family to ARF elements (AuxRE) present in the promoters of auxin-responsive genes. However, given that auxin signaling produces many…
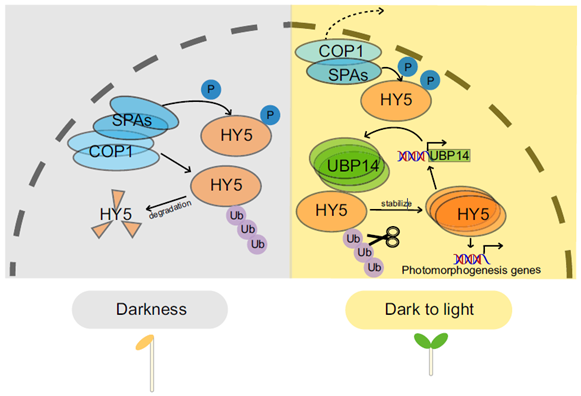
UBIQUITIN-SPECIFIC PROTEASE (UBP14) interacts with HY5 to promote photomorphogenesis under dark-to-light conditions
Plant Science Research WeeklyELONGATED HYPOCOTYL5 (HY5) is a transcription factor that regulates about one-third of Arabidopsis genes, affecting growth and development of seedlings through light and hormone signaling. UBIQUITIN-SPECIFIC PROTEASE (UBP14) is a deubiquitinating enzyme that removes ubiquitin from substrate proteins.…
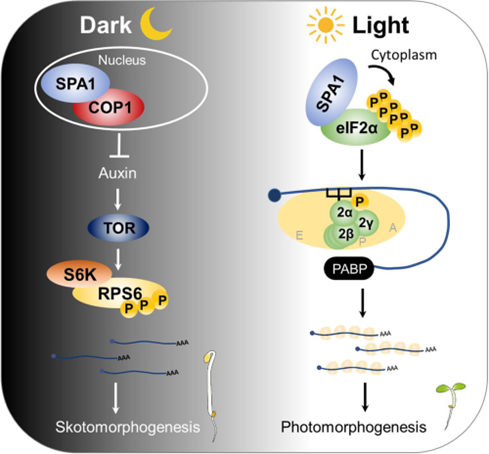
Flip the light switch for protein production
Plant Science Research WeeklyProtein synthesis, modification, and degradation are not only crucial for responding to environmental change, but they also represent significant energy costs for a plant. Energy homeostasis is highly affected by the presence of light. In young seedlings, light enhances translation during the dark-to-light…
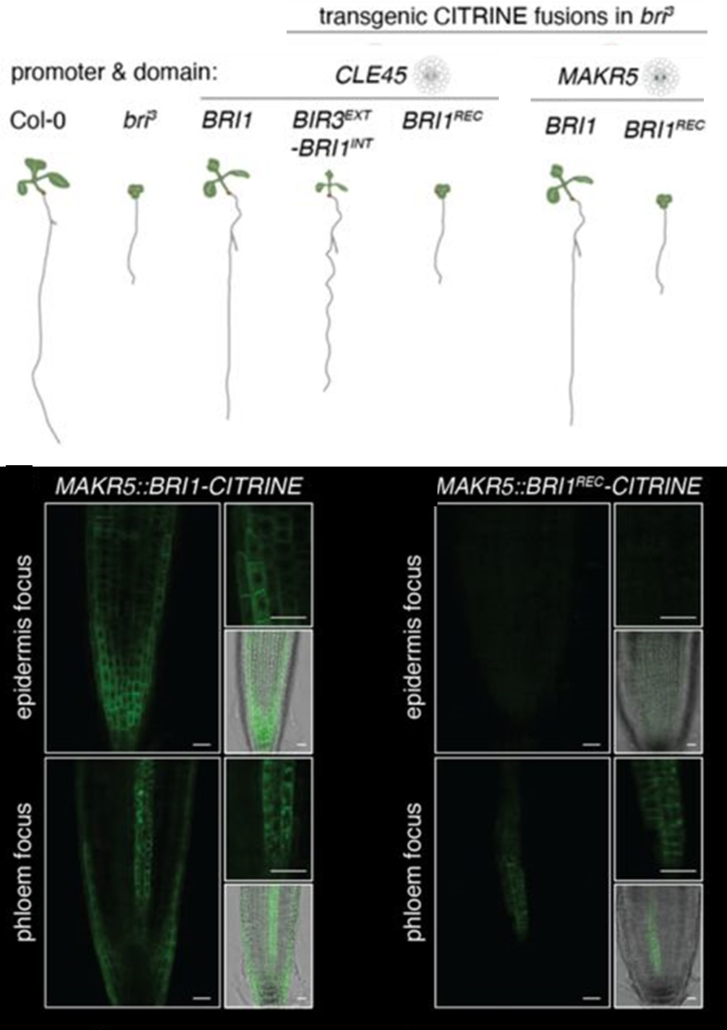
Alone or together: BRI1 signaling controls root growth in a cell-autonomous manner
Plant Science Research WeeklyRoot growth is controlled by a dense network of hormone signals working in shared and distinct root zones and cell types. Brassinosteroids (BR) control root growth through activated signaling in vascular and epidermal tissues, but it is unclear how BR signals in each tissue contribute and whether there…

