Got Rosettes? Phenotype Them Fast, Accurately, and Easily with ARADEEPOPSIS!
“Deep learning” is a buzz term that seems to be cropping up in plant biology research these days. Originally reserved, perhaps, for computer nerds rather than us biology ones, deep learning is a type of machine learning used in the field of artificial intelligence. Modeled on the human brain, deep learning uses a complex set of networks consisting of many layers (“deep”) to allow the computer to recognize (“learn”) patterns that may or may not be seen by humans, or would otherwise be too labor intensive to manually parse, in order to interpret image data. So why the recent fascination with a computational concept among us plant biologists, and why should we care?
 Well, ask yourself these questions: (1) Are you working with Arabidopsis (Arabidopsis thaliana) or other plants that exhibit a size, growth, or color phenotype? (2) Will you need to measure and analyze phenotypic differences? (3) Will you need to photograph the plants? If so, ARADEEPOPSIS, introduced by co-first authors Patrick Hüther and Niklas Schandry et al. (Hüther et al., 2020) may be in your future, and it could change your research outlook or trajectory!
Well, ask yourself these questions: (1) Are you working with Arabidopsis (Arabidopsis thaliana) or other plants that exhibit a size, growth, or color phenotype? (2) Will you need to measure and analyze phenotypic differences? (3) Will you need to photograph the plants? If so, ARADEEPOPSIS, introduced by co-first authors Patrick Hüther and Niklas Schandry et al. (Hüther et al., 2020) may be in your future, and it could change your research outlook or trajectory!
ARADEEPOPSIS (for Arabidopsis deep-learning-based optimal semantic image segmentation) is more than just a cool play on words; it is a novel, user-friendly deep-learning pipeline designed to assess phenotypes from top-view images of rosettes while eliminating a plethora of issues encountered in plant phenotyping, which is largely image based. Readily available measuring tools such as those in Fiji/ImageJ (https://imagej.net/Fiji) requires making time-consuming manual measurements that are prone to user bias and inconsistencies and are limited in scope. For measurements of rosette area, specifically, software programs (e.g., Easy Leaf Area; Easlon & Bloom 2014) exist that are designed to discriminate between “plant” and “non-plant” areas based on the green color of (most) plants. However, misidentification or lack of identification of the plant area can occur, due either to similar colors between the plant and non-plant background, as is the case when algae grows on the soil surface, or to the not-so-green color of the plant itself, as is the case for anthocyanin-rich/purplish or senescent/brownish plants. Less readily used, but improved, deep-learning methods that can extract from images both color information and structural features have been applied to plant phenotyping (Ubbens & Stavness 2017), but they require that models are generated from scratch, which is very time consuming and requires manual annotation of an extremely large number of input images for proper training of the models.
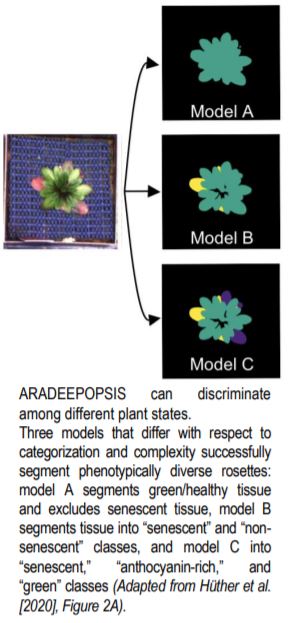
Here, using transfer learning, the authors generated over a thousand manually annotated images of rosettes in top view from 210 different Arabidopsis accessions to re-train the pre-existing DeepLabV3+ model that was originally trained on the ImageNet dataset consisting of millions of annotated images, resulting in their own models for segmentation of rosettes. ARADEEPOPSIS has various advantages to existing tools because it is more accurate and highly versatile: it can handle extremely large numbers of diverse images of varying quality and background compositions, performs a wide variety of different types of morphometric and color-index measurements of the user’s choosing, and can faithfully and accurately discriminate not just between “plant” and “non-plant” areas, but also among “green,” “anthocyanin-rich,” and “senescent” plant regions, depending on user needs (see figure). This means that plants of various sizes and shapes as well as different physiological and developmental states under various stress conditions can be assessed.
Not only did the authors test their models for segmenting using almost 150,000 rosette images from their own dataset, they also successfully validated their pipeline against published analyses that used various methods, while simultaneously demonstrating superior accuracy outputs. ARADEEPOPSIS can even be used in genome-wide association studies to determine genetic contributions to various phenotypes: as a proof of concept, the authors identified single-nucleotide polymorphisms among Arabidopsis accessions in the ANTHOCYANINLESS2 and ACCELERATED CELL DEATH 6 genes in an anthocyanin content and an early senescence analysis, respectively. Furthermore, the authors demonstrated that ARADEEPOPSIS can be applied not just to Arabidopsis but also to other species of the Brassicaceae and other plant families.
Importantly, ARADEEPOPSIS can be used on a personal laptop computer, runs on most operating systems, and does not require training in bioinformatics. Users obtain, via the Shiny web application, an interactive visual output that is well organized and includes automatically generated overlays of segmentations on original images for quality assessment. It is free and fast: depending on the number of images and the type of computer used, it is possible to obtain results in the time it takes for a quick lunch break. ARADEEPOPSIS is also customizable to other models developed by researchers that could be added to the pipeline for wider-reaching uses. While this type of (machine) learning is deep, the applications and future implications are certainly broad!
Anne C. Rea
MSU-DOE Plant Research Laboratory
Michigan State University
ORCID: 0000-0002-2996-5709
REFERENCES
Easlon, H.M. and Bloom A.J. (2014). Easy Leaf Area: Automated digital image analysis for rapid and accurate measurement of leaf area. Appl. Plant Sci. 2: 1400033.
Ubbens, J.R. and Stavness, I. (2017). Deep Plant Phenomics: A deep learning platform for complex plant phenotyping tasks. Front. Plant Sci. 8: 1190.