Comparative profiling examines roles of DNA regulatory sequences and accessible chromatin during cold stress response in grasses
Physical access to regulatory DNA, including cis-regulatory sequences found within proximal promoters and distal enhancer elements, is a vital property of chromatin. In turn, their access is determined by nucleosome occupancy and post-translational modification of histone proteins. A continuum of chromatin accessibility states from closed to permissive and open chromatin is a critical determinant of chromatin’s regulatory capacity in modulating gene expression. Transcription factors (TFs) can dynamically regulate local access to DNA by modulating nucleosome occupancy; in turn, the cell/tissue-specific chromatin landscape affects TF binding. Recent identification of chromatin signatures of cis-regulatory elements across angiosperms provides a strong foundation for understanding TFs and their interaction with the chromatin environment in modulating gene expression (Lu et al. 2019).
 In new work, Han et al. (2020) attempt to dissect the dynamics of the chromatin landscape and regulatory elements in response to cold stress. The authors performed a comprehensive analysis of active regulatory DNA through DNase I-hypersensitive site (DHS) mapping of control and cold-stressed samples from three different tissue types – root, stem, and leaves – in three grass species – Brachypodium distachyon, Setaria italica, and Sorghum bicolor. Incorporating multiple datasets such as DNase-seq, RNA-seq, and Hi-C, the authors identified tissue- and species-specific patterns of open chromatin in response to cold stress. Consistent expression of a GFP-reporter gene driven by predicted B. distachyon leaf DHSs in protoplast-based transient transformation provided experimental validation of promoter and enhancer functions of a subset of the identified DHSs.
In new work, Han et al. (2020) attempt to dissect the dynamics of the chromatin landscape and regulatory elements in response to cold stress. The authors performed a comprehensive analysis of active regulatory DNA through DNase I-hypersensitive site (DHS) mapping of control and cold-stressed samples from three different tissue types – root, stem, and leaves – in three grass species – Brachypodium distachyon, Setaria italica, and Sorghum bicolor. Incorporating multiple datasets such as DNase-seq, RNA-seq, and Hi-C, the authors identified tissue- and species-specific patterns of open chromatin in response to cold stress. Consistent expression of a GFP-reporter gene driven by predicted B. distachyon leaf DHSs in protoplast-based transient transformation provided experimental validation of promoter and enhancer functions of a subset of the identified DHSs.
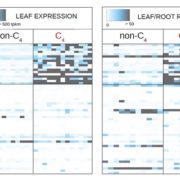
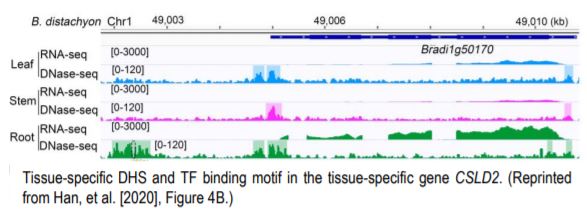
Many more highly expressed genes contained genic DHSs than moderate to lowly expressed genes, and highly expressed genes on average had more DHSs in all three grass species. Genes with more than four DHSs tended to be tissue-specific and correlated with a higher dynamic range of expression. Consistent with previous reports, including one with the same three species, the authors show low conservation of accessible chromatin (Maher et al., 2018; Burgess et al., 2019). However, recent work from Lu et al. (2019) show that more than half of the accessible chromatin regions are conserved between closely related pairs of species, maize-sorghum, and soybean-phaseolus. A significant proportion of tissue-specific genes showed a correlation with tissue-specific DHS, as far as 50 kb, suggesting potential distal regulation of tissue-specific gene expression. This long-range interaction was confirmed in S. italica and S. bicolor using Hi-C data from a previous study. Tissue-specific DHSs were shown to regulate the expression of tissue-specific genes. For example, cellulose synthase-like gene CSLD2 (Bradi1g50170), and its orthologs showed root-specific gene expression. Of the two DHSs upstream of the gene, one was shared across three tissues. At the same time, the other DHS was root-specific (given their experimental resolution) and present in all three species, containing MYC-like sequence motif, CATGTG, potentially targeted by NAC TFs affecting the differential expression of CSLD2 in roots (Figure).
A significant proportion of cold-induced DHSs was adjacent to genes differentially expressed between control and cold-treated samples. Although they did not find any common cold-induced DHS shared across the three species or the three tissue types in each species, cold-induced TF-binding motifs showed higher conservation. These conserved motifs suggest similar transcriptional regulation in different cell types controlled by common TFs across species in specific biological functions. Cold-induced DHSs were almost exclusively tissue-specific, suggesting that conservation of cold stress response in grasses is mostly tissue-specific. The authors identified 17 TFs shared across all three species in the three tissues tested, including known regulators of cold stress-responsive genes.
Han et al. provide a dynamic view of accessible chromatin and transcriptional regulatory networks in response to an important environmental challenge. Accessible chromatin is variable in number and location across species (Maher et al., 2018), and a complete understanding of core regulatory mechanisms behind complex traits like stress response is not feasible merely by looking at accessible chromatin. However, despite low conservation of accessible chromatin regions, TF binding motifs are conserved, highlighting shared transcriptional regulation in different tissue types across species in specific biological functions (Maher et al., 2018; Reynoso et al., 2019).
Sunil K. Kenchanmane Raju
Department of Plant Biology,
Michigan State University
ORCID ID: 0000-0001-8960-094X
REFERENCES
Lu, Z., Marand, A. P., Ricci, W. A., Ethridge, C. L., Zhang, X., & Schmitz, R. J. (2019). The prevalence, evolution and chromatin signatures of plant regulatory elements. Nature Plants, 5: 1250-1259.
Han, J., Wang, P., Wang, Q., Lin, Q., Chen, Z., Yu, G., … & Tang, H. (2020). Genome-wide Characterization of DNase I-hypersensitive Sites and Cold Response Regulatory Landscapes in Grasses. Plant Cell. DOI: https://doi.org/10.1105/tpc.19.00716
Maher, K. A., Bajic, M., Kajala, K., Reynoso, M., Pauluzzi, G., West, D. A., … & Queitsch, C. (2018). Profiling of accessible chromatin regions across multiple plant species and cell types reveals common gene regulatory principles and new control modules. Plant Cell, 30: 15-36.
Burgess, S. J., Reyna-Llorens, I., Stevenson, S. R., Singh, P., Jaeger, K., & Hibberd, J. M. (2019). Genome-wide transcription factor binding in leaves from C3 and C4 grasses. Plant Cell, 31: 2297-2314.
Reynoso, M. A., Kajala, K., Bajic, M., West, D. A., Pauluzzi, G., Yao, A. I., … & Sinha, N. (2019). Evolutionary flexibility in flooding response circuitry in angiosperms. Science, 365: 1291-1295.