Mix, Match, and Maize: A Synthetic System for Maize Nuclear Auxin Response Circuits
Dhineshkumar Thiruppathi 1,2
Donald Danforth Plant Science Center,
Saint Louis, Missouri 63132
1Lead author
2Author for contact: [email protected]
The phytohormone auxin plays a major part in nearly every plant process, including growth and development. Auxin performs these functions, in part, by eliciting context-specific nuclear responses mediated by a signaling network (Leyser, 2018). Each core component of this network belongs to a large gene family, varies in copy number, and generates diversity in local auxin responses (Mutte et al., 2018). As such, the evolution of complex body plans in land plants is linked to gene duplication and divergence within this network, highlighting the need for functional analysis of auxin circuitry components across multiple species. A mechanistic understanding of auxin circuitry also has the potential to allow rational engineering of novel/desirable traits in agriculturally important plants. In this issue of Plant Physiology Ramos Báez et al. (2020) approach the challenge of auxin network analysis, focusing on the response circuitry of maize (Zea mays) inflorescence development.
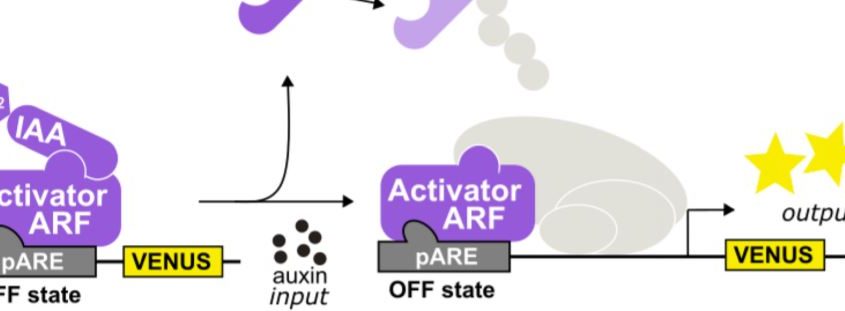
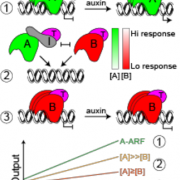
 A general model for nuclear auxin responses integrates four major families of proteins. Under low auxin, AUXIN/INDOLE-3-ACETIC-ACID INDUCIBLE (Aux/IAA) repressor proteins interact with TOPLESS (TPL) co-repressor proteins and repress AUXIN RESPONSE FACTOR (ARF) transcription factor-mediated gene transcription. Upon increased auxin, TRANSPORT INHIBITOR RESPONSE1 (TIR1)/AUXIN SIGNALING F-BOX (AFB) receptor family members perceive auxin, permitting Aux/IAA interaction. The TIR1/AFB-auxin-Aux/IAA complex leads to Aux/IAA protein degradation, freeing ARFs to regulate auxin-responsive gene expression (Lavy and Estelle, 2016). Developed on this knowledge, a yeast synthetic system known as ARCSc (Auxin Response Circuit recapitulated in Saccharomyces cerevisiae) allowed studies on the dynamic capabilities of auxin signaling in isolation. Importantly, this system revealed extensive tunability of the auxin response by specific configurations of the auxin circuit family members, for example AtARCSc in Arabidopsis (Arabidopsis thaliana; Pierre-Jerome et al., 2014). Ramos Báez and colleagues adapted this platform to characterize the gene families of the maize auxin response circuits in flowering. They created a ZmARCSc degradation module to measure differences in auxin-induced degradation rates of Aux/IAAs by the yeast ubiquitin machinery, and they created a ZmARCSc repression module to measure auxin-induced derepression of an Aux/IAA-TPL fusion, each with a fluorescent marker for quantification.
A general model for nuclear auxin responses integrates four major families of proteins. Under low auxin, AUXIN/INDOLE-3-ACETIC-ACID INDUCIBLE (Aux/IAA) repressor proteins interact with TOPLESS (TPL) co-repressor proteins and repress AUXIN RESPONSE FACTOR (ARF) transcription factor-mediated gene transcription. Upon increased auxin, TRANSPORT INHIBITOR RESPONSE1 (TIR1)/AUXIN SIGNALING F-BOX (AFB) receptor family members perceive auxin, permitting Aux/IAA interaction. The TIR1/AFB-auxin-Aux/IAA complex leads to Aux/IAA protein degradation, freeing ARFs to regulate auxin-responsive gene expression (Lavy and Estelle, 2016). Developed on this knowledge, a yeast synthetic system known as ARCSc (Auxin Response Circuit recapitulated in Saccharomyces cerevisiae) allowed studies on the dynamic capabilities of auxin signaling in isolation. Importantly, this system revealed extensive tunability of the auxin response by specific configurations of the auxin circuit family members, for example AtARCSc in Arabidopsis (Arabidopsis thaliana; Pierre-Jerome et al., 2014). Ramos Báez and colleagues adapted this platform to characterize the gene families of the maize auxin response circuits in flowering. They created a ZmARCSc degradation module to measure differences in auxin-induced degradation rates of Aux/IAAs by the yeast ubiquitin machinery, and they created a ZmARCSc repression module to measure auxin-induced derepression of an Aux/IAA-TPL fusion, each with a fluorescent marker for quantification.
The authors trialed this system using maize Aux/IAAs (ZmIAAs) that were known to be expressed in developing inflorescences through RNA in situ hybridization. They found that the inflorescence-expressed ZmIAAs are functional in auxin degradation and repression modules including BARREN INFLORESCENCE1 (BIF1), a ZmIAA known to be essential for inflorescence development (Galli et al., 2015). In particular, these ZmIAAs exhibited differential but more rapid degradation rates, as measured YFP signals by flow cytometry in ARCSc degradation assays with AtAFB2 than those with AtTIR1. In ARCSc repression modules with AtAFB2, the ZmIAAs fused to RAMOSA1 ENHANCER LOCUS2 (REL2), an inflorescence-specific maize TPL homolog also exhibited variations in their ability to repress and, upon auxin treatment, derepress AtARF19 induction of pARE, as measured VENUS signals by flow cytometry.
Expanding their screen, Ramos Báez et al., (2020) found that a functional ARC featuring the ZmAFB2/3 b1 receptor was more sensitive to auxin than a circuit with its ortholog AtAFB2, previously the fastest acting known Arabidopsis receptor. ZmAFB2/3 b1 belongs to a clade of AFB2/3-like maize auxin receptors that are highly expressed in developing inflorescence meristems (Eveland et al., 2014). The authors paired ZmAFB2/3 b1 in their ARC auxin repression module with various ZmIAAs and found different degradation kinetics for each of the ZmIAAs. In particular, the inflorescence development regulator BIF1 was degraded faster than ZmIAA8 and ZmIAA12. Such patterns of degradation were even faster than that observed with AtAFB2 co-expression. They concluded that ZmAFB2/3 b1 has a higher basal activity than AtAFB2, a difference linked to a diverging residue essential for auxin sensitivity.
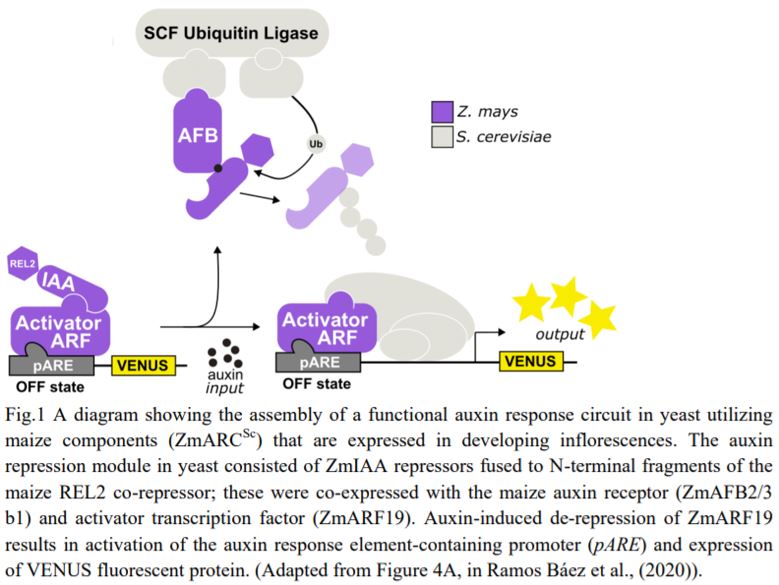
Finally, the authors expressed the functional maize receptors, repressors, and co-repressors with a maize ARF to reconstitute an entire ZmARCSc maize auxin response circuit (Fig. 1) in yeast. They generated ZmARCSc variants expressing ZmIAAs together with ZmARF27, an ARF expressed strongly in immature tassel and orthologues to AtARF19 used in the AtARCSc, and monitored the ZmARF27-activated pARE response. Not surprisingly, the various ZmIAAs repressed ZmARF27 to varying degrees and derepressed it upon auxin treatment. Significantly, however, they found that the ZmARCSc circuit with the ZmAFB2/3 b1 responded to lower doses of auxin than that with AtAFB2, indicating that the maize ARC is exquisitely sensitive to auxin hormone levels.
Overall, Ramos Báez et al., (2020) provide a synthetic mix-and-match platform for systemic examination of auxin circuitry components of maize. Their studies set a landmark for comparative studies across distantly related and phenotypically complex plants with sequenced genomes. The analysis of interspecific variation in auxin sensitivity that they demonstrate extends previous work on the evolution of auxin pathway by focusing on the functional dynamics at the level of the component proteins. It remains to be seen whether the minimal assembly of such signaling modules mimics the networks found in planta. Nonetheless, this study serves as a stepping stone for future investigations that will generate hypotheses about evolution of ARC variants in plants. Equally exciting, it is likely that a similar strategy can be applied in analyzing signaling pathways that are regulated by other phytohormones that use derepression mechanisms similar to that of auxin such as gibberellic acid, jasmonic acid, and strigolactone (Blázquez et al., 2020).
LITERATURE CITED
Blázquez MA, Nelson DC, Weijers D (2020) Evolution of Plant Hormone Response Pathways. Annu Rev Plant Biol. doi: 10.1146/annurev-arplant-050718-100309
Eveland AL, Goldshmidt A, Pautler M, Morohashi K, Liseron-Monfils C, Lewis MW, Kumari S, Hirag S, Yang F, Unger-Wallace E, et al (2014) Regulatory modules controlling maize inflorescence architecture. Genome Res. doi: 10.1101/gr.166397.113
Galli M, Liu Q, Moss BL, Malcomber S, Li W, Gaines C, Federici S, Roshkovan J, Meeley R, Nemhauser JL, et al (2015) Auxin signaling modules regulate maize inflorescence architecture. Proc Natl Acad Sci U S A. doi: 10.1073/pnas.1516473112
Lavy M, Estelle M (2016) Mechanisms of auxin signaling. Dev. doi: 10.1242/dev.131870
Leyser O (2018) Auxin signaling. Plant Physiol. doi: 10.1104/pp.17.00765
Mutte SK, Kato H, Rothfels C, Melkonian M, Wong GKS, Weijers D (2018) Origin and evolution of the nuclear auxin response system. Elife. doi: 10.7554/eLife.33399
Pierre-Jerome E, Jang SS, Havens KA, Nemhauser JL, Klavins E (2014) Recapitulation of the forward nuclear auxin response pathway in yeast. Proc Natl Acad Sci U S A. doi: 10.1073/pnas.1324147111