The Epigenetics of Leaf Senescence
Liu et al. explore the mechanism underlying the epigenetic reprograming of senescence-associated gene expression during leaf senescence. Plant Cell https://doi.org/10.1105/tpc.18.00693
By Peng Liu, Jing Bo Jin, and Xiaofeng Cao, Chinese Academy of Sciences, Beijing, China.
Background: Leaf senescence is the highly ordered final stage of leaf development. During leaf senescence, chlorophylls, nucleic acids, lipids, proteins, and other macromolecules are degraded, and the released nutrients are recycled to growing and storage organs. The transition from maturity to senescence in leaves is associated with dramatic changes in the expression of thousands of senescence-associated genes (SAGs). The induction of SAG transcription during leaf senescence is largely correlated with increased trimethylation of histone H3 at lysine 4 (H3K4me3, a mark of actively transcribed chromatin) levels at the chromatin regions of SAGs. However, how H3K4me3 levels at the SAGs are regulated in an age-dependent manner remains largely unknown.
Question: H3K4me3 levels are regulated by histone methyltransferases (writers) and histone demethylases (erasers). We wanted to know if histone demethylase(s) regulates the age-dependent induction of H3K4me3 levels of the SAGs. We tested this by determining the leaf senescence phenotypes of known or putative H3K4 histone demethylase loss-of-function mutants in Arabidopsis thaliana.
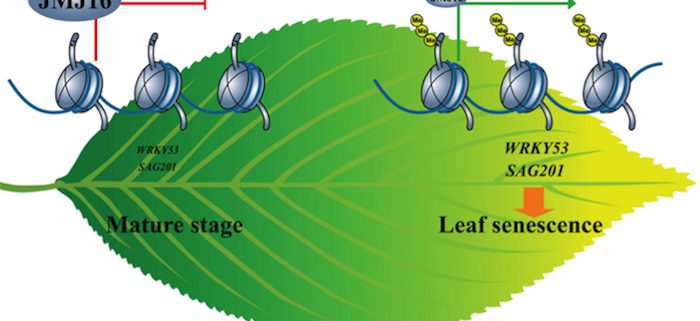
Findings: We found that mutations in JMJ16, encoding an Arabidopsis JmjC-domain containing protein, caused early leaf senescence. JMJ16 exhibits H3K4 demethylase activity and negatively regulates age-triggered leaf senescence through its enzymatic activity. In mature leaves (prior to leaf senescence), JMJ16 associates with WRKY53 and SAG201, two positive regulators of leaf senescence, and represses their precocious expression by reducing H3K4me3 levels at these loci, thereby preventing precocious activation of the senescence program. During the transition to leaf senescence, JMJ16 protein levels are substantially decrease. Consequently, H3K4me3 levels and WRKY53 and SAG201 transcript levels increase, and thereby initiating the senescence program.
Next steps: Currently, we do not understand how JMJ16 protein levels are downregulated in an age-dependent manner. It would be interesting to determine if post-translational modifications, such as ubiquitination, regulate JMJ16 protein stability during leaf aging. Our study suggests that H3K4 methyltransferase(s) may also regulates age-dependent activation of SAGs, but this notion remains to be investigated.
Peng Liu, Shuaibin Zhang, Bing Zhou, Xi Luo, Xiao Feng Zhou, Bin Cai, Yin Hua Jin, De Niu, Jinxing Lin, Xiaofeng Cao, and Jing Bo Jin. (2019). The Histone H3K4 Demethylase JMJ16 Represses Leaf Senescence in Arabidopsis. Plant Cell; DOI: https://doi.org/10.1105/tpc.18.00693
Key words: Senescence, histone, epigenetics, histone demethylase, leaf