Similarity between soybean and Arabidopsis seed methylomes and loss of non-CG methylation does not affect seed development
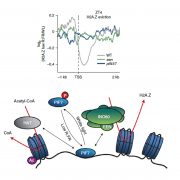
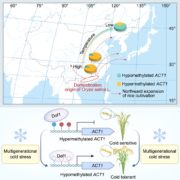
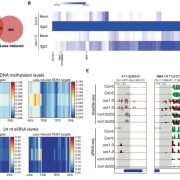
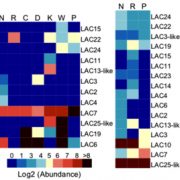
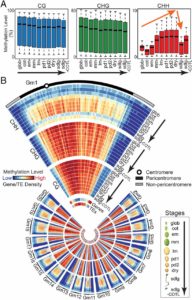 Different parts of the seed have distinct genetic origins and functions. The seed coat is maternally derived from ovule integuments, whereas the embryo descends from the fertilized egg and the endosperm from the central cell. Lin et al. profiled the methylome landscape during seed development and germination in Arabidopsis and soybean using laser capture microdissection and whole-genome bisulfite sequencing. DNA methylation architecture during seed development and germination appears to be conserved between soybean and Arabidopsis despite ~90 MY divergence. They show that CHH methylation drastically increases during seed development, targeting all classes of transposable elements (TEs). Later during germination, both the bulk CHH methylation levels and the number of mCHH sites decreases. In contrast, CG and CHG methylation levels remain more or less constant during seed development. To understand the role of DNA methylation changes on the expression of genes known to be important for seed development and germination in both soybean and Arabidopsis, the authors characterized the methylation patterns and expression levels of ~75 genes. Most of these genes were found in regions of very low DNA methylation, which the authors termed ‘demethylated valleys’. The surrounding methylation levels of these genes didn’t change during seed development, despite changes in gene expression. The authors suggest that the regulation of seed development genes results primarily from transcriptional events that are independent of DNA methylation changes. Furthermore, Arabidopsis mutants lacking CHG and CHH methylation develop and germinate normally and don’t show much change to the transcriptome, except for the increased expression of transposons. This suggests that CHH hypermethylation during seed development may be a failsafe mechanism to avoid deleterious effects of TE proliferation. PNAS
Different parts of the seed have distinct genetic origins and functions. The seed coat is maternally derived from ovule integuments, whereas the embryo descends from the fertilized egg and the endosperm from the central cell. Lin et al. profiled the methylome landscape during seed development and germination in Arabidopsis and soybean using laser capture microdissection and whole-genome bisulfite sequencing. DNA methylation architecture during seed development and germination appears to be conserved between soybean and Arabidopsis despite ~90 MY divergence. They show that CHH methylation drastically increases during seed development, targeting all classes of transposable elements (TEs). Later during germination, both the bulk CHH methylation levels and the number of mCHH sites decreases. In contrast, CG and CHG methylation levels remain more or less constant during seed development. To understand the role of DNA methylation changes on the expression of genes known to be important for seed development and germination in both soybean and Arabidopsis, the authors characterized the methylation patterns and expression levels of ~75 genes. Most of these genes were found in regions of very low DNA methylation, which the authors termed ‘demethylated valleys’. The surrounding methylation levels of these genes didn’t change during seed development, despite changes in gene expression. The authors suggest that the regulation of seed development genes results primarily from transcriptional events that are independent of DNA methylation changes. Furthermore, Arabidopsis mutants lacking CHG and CHH methylation develop and germinate normally and don’t show much change to the transcriptome, except for the increased expression of transposons. This suggests that CHH hypermethylation during seed development may be a failsafe mechanism to avoid deleterious effects of TE proliferation. PNAS