Single nucleus analysis of Arabidopsis seeds reveals new cell types and imprinting dynamics (bioRxiv)
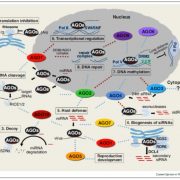
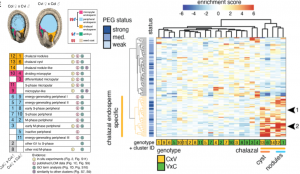 Arabidopsis seeds consist of various tissue like seed coat, embryo, and endosperm. The endosperm provides the nutrient supplies to the growing embryo and has three domains namely, micropylar (surrounding embryo), chalazal (opposite end of the seed), and peripheral (in between micropylar and chalazal) domain. Even though there is transcriptomic data available for these different domains of endosperm, it is not yet known whether there is cell-type heterogeneity within these domains. In this paper Picard et al. carried out single nucleus RNA-sequencing (snRNA-Seq) on F1 seeds from a reciprocal cross between the Columbia (Col-0) and Cape Verde Islands (Cvi-0) to identify cell-type and imprinting heterogeneity in the endosperm. Using data from snRNA-Seq, the authors identified two distinct cell types in the chalazal domain based on gene expression analysis and cell cycle phase. Using the allelic gene expression data from their reciprocal crosses the authors found that there is almost no heterogeneity in the maternally expressed genes whereas paternally expressed genes show biased expression in the chalazal endosperm. Thus, this study identifies a cell-type and imprinting heterogeneity and provides a unique opportunity to compare gene expression, cell cycle, and imprinting in seemingly complex endosperm domains. Summary by (Vijaya Batthula @Vijaya_Batthula). bioRxiv. 10.1101/2020.08.25.267476.
Arabidopsis seeds consist of various tissue like seed coat, embryo, and endosperm. The endosperm provides the nutrient supplies to the growing embryo and has three domains namely, micropylar (surrounding embryo), chalazal (opposite end of the seed), and peripheral (in between micropylar and chalazal) domain. Even though there is transcriptomic data available for these different domains of endosperm, it is not yet known whether there is cell-type heterogeneity within these domains. In this paper Picard et al. carried out single nucleus RNA-sequencing (snRNA-Seq) on F1 seeds from a reciprocal cross between the Columbia (Col-0) and Cape Verde Islands (Cvi-0) to identify cell-type and imprinting heterogeneity in the endosperm. Using data from snRNA-Seq, the authors identified two distinct cell types in the chalazal domain based on gene expression analysis and cell cycle phase. Using the allelic gene expression data from their reciprocal crosses the authors found that there is almost no heterogeneity in the maternally expressed genes whereas paternally expressed genes show biased expression in the chalazal endosperm. Thus, this study identifies a cell-type and imprinting heterogeneity and provides a unique opportunity to compare gene expression, cell cycle, and imprinting in seemingly complex endosperm domains. Summary by (Vijaya Batthula @Vijaya_Batthula). bioRxiv. 10.1101/2020.08.25.267476.