Real time quantitative imaging of transcriptional activity at the single cell level (bioRxiv)
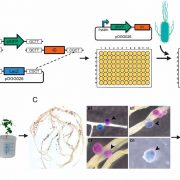
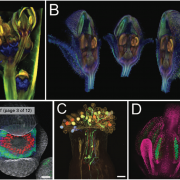
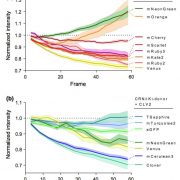
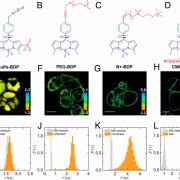
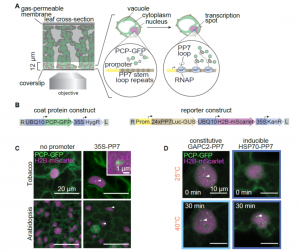 How plants transcriptionally operate developmental programs and responses to stress in time and space has been an important question in plant biology. Fluorescent protein reporters are commonly used to address this question, but their performance is limited at short timescales (<30 min) before the proteins get matured. Also, expression of fluorescent proteins can be influenced by various processes before and after maturation (e.g., RNA transport and protein degradation). To overcome these limitations, Alamos et al. implemented an mRNA fluorescent-tagging approach in Nicotiana benthamiana and Arabidopsis thaliana. In this system, the promoter of a gene is tagged with the PP7 (or MS2) viral DNA sequence that when transcribed accumulates fluorescent proteins already expressed in the nucleus. The signal can be immediately detected as a fluorescent spot, and the signal intensity reports the absolute count of RNA polymerase molecules actively transcribing the target gene. The authors generated stably transformed lines of Arabidopsis with construct targeting the heat shock protein HSP101 to monitor plant responses to heat stress. They found that only a fraction of cells was transcriptionally responsive to the stress, and among responsive cells, instantaneous transcription events were detected in a heterogeneous manner. The tissue-wide transcription rate of the heat-responsive gene was controlled by a switch-like mechanism where heat stress induces the probability that individual cells engage in transcription, whereas the transcription level of already actively transcribing cells remained stable. Finally, they showed that the cellular heterogeneity in the transcriptional response to heat shock is mainly explained by stochastic processes at the level of each allele rather than cells having a different chemical composition. This approach opens the door for single cell interrogation of transcription variability and dynamics in plants. (Summary by Tatsuya Nobori @nobolly) bioRxiv 10.1101/2020.08.30.274621
How plants transcriptionally operate developmental programs and responses to stress in time and space has been an important question in plant biology. Fluorescent protein reporters are commonly used to address this question, but their performance is limited at short timescales (<30 min) before the proteins get matured. Also, expression of fluorescent proteins can be influenced by various processes before and after maturation (e.g., RNA transport and protein degradation). To overcome these limitations, Alamos et al. implemented an mRNA fluorescent-tagging approach in Nicotiana benthamiana and Arabidopsis thaliana. In this system, the promoter of a gene is tagged with the PP7 (or MS2) viral DNA sequence that when transcribed accumulates fluorescent proteins already expressed in the nucleus. The signal can be immediately detected as a fluorescent spot, and the signal intensity reports the absolute count of RNA polymerase molecules actively transcribing the target gene. The authors generated stably transformed lines of Arabidopsis with construct targeting the heat shock protein HSP101 to monitor plant responses to heat stress. They found that only a fraction of cells was transcriptionally responsive to the stress, and among responsive cells, instantaneous transcription events were detected in a heterogeneous manner. The tissue-wide transcription rate of the heat-responsive gene was controlled by a switch-like mechanism where heat stress induces the probability that individual cells engage in transcription, whereas the transcription level of already actively transcribing cells remained stable. Finally, they showed that the cellular heterogeneity in the transcriptional response to heat shock is mainly explained by stochastic processes at the level of each allele rather than cells having a different chemical composition. This approach opens the door for single cell interrogation of transcription variability and dynamics in plants. (Summary by Tatsuya Nobori @nobolly) bioRxiv 10.1101/2020.08.30.274621