Chen Mingsheng’s research team found an evolutionary trend of genes fleeing the centromere region
Article source: http://theworldseeds.cn/index.php?p=152804 (Translated by Google Translate)
The centromere and its surroundings are the fastest-evolving and most complex areas of the plant genome. The centromere and near centromere regions not only undergo rapid sequence changes and structural remodeling, but also have transcriptionally active genes and are also hotspots for the origin of new genes.
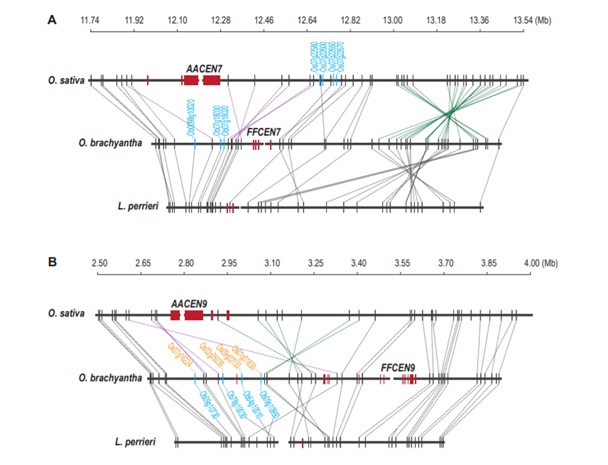
Chen Mingsheng, a research group of the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, completed the whole genome sequencing of short anther wild rice (Oryza brachyantha), and used BAC sequencing and physical maps to improve the sequence of the near centromere region of the twelve chromosome centromeres of Oryza brachyantha. Based on this, a comparative genomics study on the centromere region of Oryza and its relatives was carried out. The study found that the Oryza brachyantha was independently selected and adapted to the specific centromere sequence; the inversion of the near centromere was the main way to move the centromere position; the position of the Oryza brachyantha chromosome 12 centromere movement is a typical centromere relocation phenomenon. The new centromere originates from a segmental repeat of the old centromere region; the gene in the centromere region or its periphery is selectively deleted by gene duplication, and the gene escapes. The evolutionary trend of the centromere environment, which is derived from the adverse effects of the genetic and epigenetic environment on the gene on the one hand, and may also be related to the expansion of the centromere on the other hand. The region has formed a large number of new genes during the recent evolution.
The study was published online July 2, 2018 in The Plant Cell ( DOI:10.1105/tpc.18.00163). Dr. Liao Yi and Dr. Zhang Xuemei from the Chen Mingsheng Research Group are co-first authors of the paper. The study was funded by the National Natural Science Foundation of China.
In case any of that was garbled in translation, here is the abstract of the paper
ABSTRACT
Centromeres are dynamic chromosomal regions and the genetic and epigenetic environment of the centromere is often regarded as oppressive to protein-coding genes. Here we used comparative genomic and phylogenomic approaches to study the evolution of centromeres and centromere-linked genes in the genus Oryza. We report a 12.4 Mb high-quality BAC40 based pericentromeric assembly for Oryza brachyantha, which diverged from cultivated rice (Oryza sativa) ~15 Mya (million years ago). The synteny analyses reveal seven medium (>50 kb) pericentric inversions in O. sativa and ten in O. brachyantha. Of 42 these inversions, three resulted in centromere movement (Chr1, Chr7, and Chr9). Additionally, we identified a potential centromere-repositioning event, in which the ancestral centromere on chromosome 12 in Oryza brachyantha jumped ~ 400 kb away, possibly mediated by a duplicated transposition event (>28 kb). More strikingly, we observed an excess of syntenic gene loss at and near the centromeric regions (P < 2.2×10-16). Most (33/47) of the missing genes moved to other genomic regions; therefore such excess could be explained by the selective loss of the copy in or near centromeric regions after gene duplication. The pattern of gene loss immediately adjacent to centromeric regions suggests centromere chromatin dynamics (e.g. spreading or micro-repositioning) may drive such gene loss.