The subgenomes of polyploid plants evolve at different rates (Plant Cell)
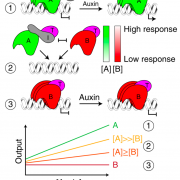
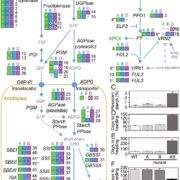
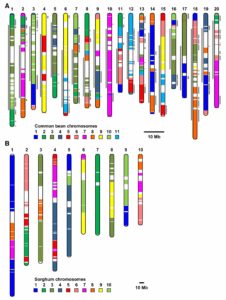 Polyploidy can result either through genome doubling followed by intraspecific crosses, or when two genomes from independent species hybridize, leading to autopolyploidy, or allopolyploidy, respectively. Polyploidy is an important evolutionary tool; the resultant redundancy from the duplication of genes allows for neofunctionalisation or subfunctionalisation to occur, which can lead to reproductive isolation, facilitating speciation and evolution. The evolutionary fate of subgenomes is dependent on whether the polyploid is the result of autopolyploidy or allopolyploidy, and how different the initial genomes where from one another. A recent study by Zhao et al. investigated the factors influencing the evolution of duplicated gene pairs in polyploids by comparing genomic and epigenomic differences in the genomes of maize and soybean. Maize likely resulted from an allopolyploidy event, and because of the initial differences between the two progenitor genomes, one subgenome exhibits dominance in gene expression and selective constraints over the other, influencing how the genome changes over time. Soybean, conversely, shows little evidence for subgenome dominance, and was, therefore, likely the result of an autopolyploidy, or an allopolyploidy from two closely related species, resulting in the soybean genome retaining the majority of duplicate genes. This study sheds light on the factors driving genome evolution, and what effects subgenome dominance can have over long time periods. (Summary by Danielle Roodt Prinsloo) Plant Cell 10.1105/tpc.17.00595
Polyploidy can result either through genome doubling followed by intraspecific crosses, or when two genomes from independent species hybridize, leading to autopolyploidy, or allopolyploidy, respectively. Polyploidy is an important evolutionary tool; the resultant redundancy from the duplication of genes allows for neofunctionalisation or subfunctionalisation to occur, which can lead to reproductive isolation, facilitating speciation and evolution. The evolutionary fate of subgenomes is dependent on whether the polyploid is the result of autopolyploidy or allopolyploidy, and how different the initial genomes where from one another. A recent study by Zhao et al. investigated the factors influencing the evolution of duplicated gene pairs in polyploids by comparing genomic and epigenomic differences in the genomes of maize and soybean. Maize likely resulted from an allopolyploidy event, and because of the initial differences between the two progenitor genomes, one subgenome exhibits dominance in gene expression and selective constraints over the other, influencing how the genome changes over time. Soybean, conversely, shows little evidence for subgenome dominance, and was, therefore, likely the result of an autopolyploidy, or an allopolyploidy from two closely related species, resulting in the soybean genome retaining the majority of duplicate genes. This study sheds light on the factors driving genome evolution, and what effects subgenome dominance can have over long time periods. (Summary by Danielle Roodt Prinsloo) Plant Cell 10.1105/tpc.17.00595