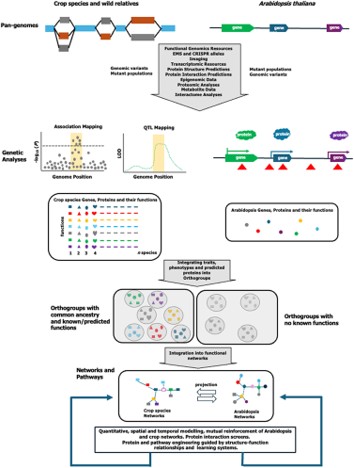
Review: Gene discovery, from Arabidopsis to crops
Plant Science Research WeeklyThis year marks the 25th anniversary of the publication of the first genome sequence of a plant, Arabidopsis thaliana, (see https://doi.org/10.1038/35048692). As this review by Bevan et al. observes, this exciting accomplishment was met with some skepticism by those who felt that it was not likely to…
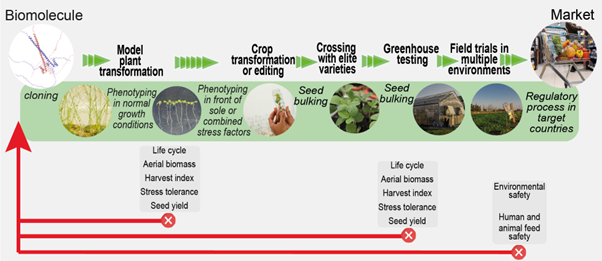
Review: Translating research from Arabidopsis to crops
Plant Science Research WeeklyThis year marks the 25th anniversary of the publication of the Arabidopsis genome, a milestone that lives large in those of us who experienced it and eagerly envisioned how this new knowledge would be used. The May 2025 issue of The Plant Cell focuses on how research on this little model plant has translated…
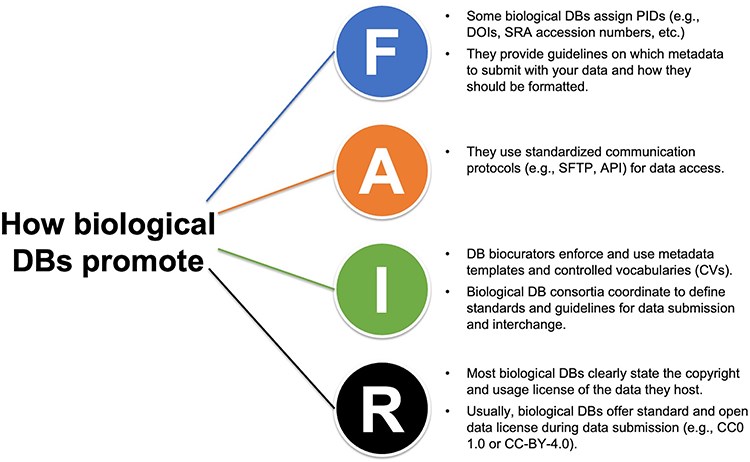
Training scientists to make their data FAIR
Plant Science Research WeeklyI’m sure many of you have experienced frustration when trying to access an intriguing dataset that either doesn’t exist, isn’t open, or is set up in an impossibly unintuitive manner. Part of that problem stems from a lack of training of early-career scientists in how to make their data findable,…
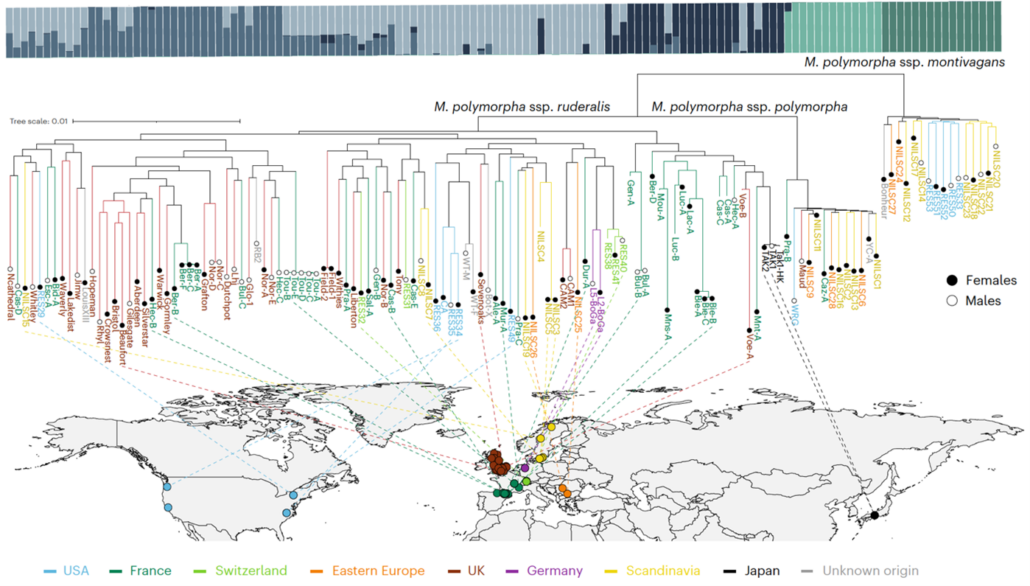
From water to land: What bryophytes reveal about plant evolution and adaptations
Plant Science Research WeeklyThe transition of plants from aquatic to terrestrial environments occurred approximately 400 million years ago, leading to the diversification of two major lineages: tracheophytes (vascular plants) and bryophytes (non-vascular plants). While most studies on plant adaptation to environmental stressors…
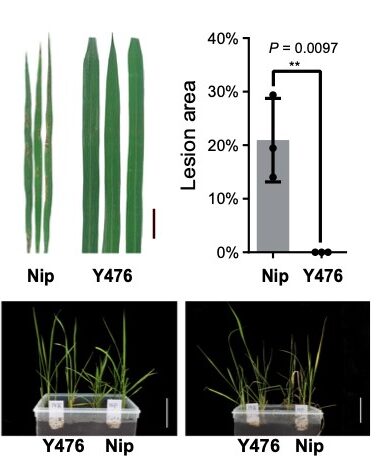
Crossing the gap: Decrypting the genome facilitates gene identification in wild rice
Plant Science Research WeeklyDomesticated crops provide a reliable food source but generally have little genetic diversity to cope with environmental fluctuations. By decrypting the genome of wild species such as the wild rice Oryza rufipogon, we may identify additional genetic variation useful for breeding process. However, the…
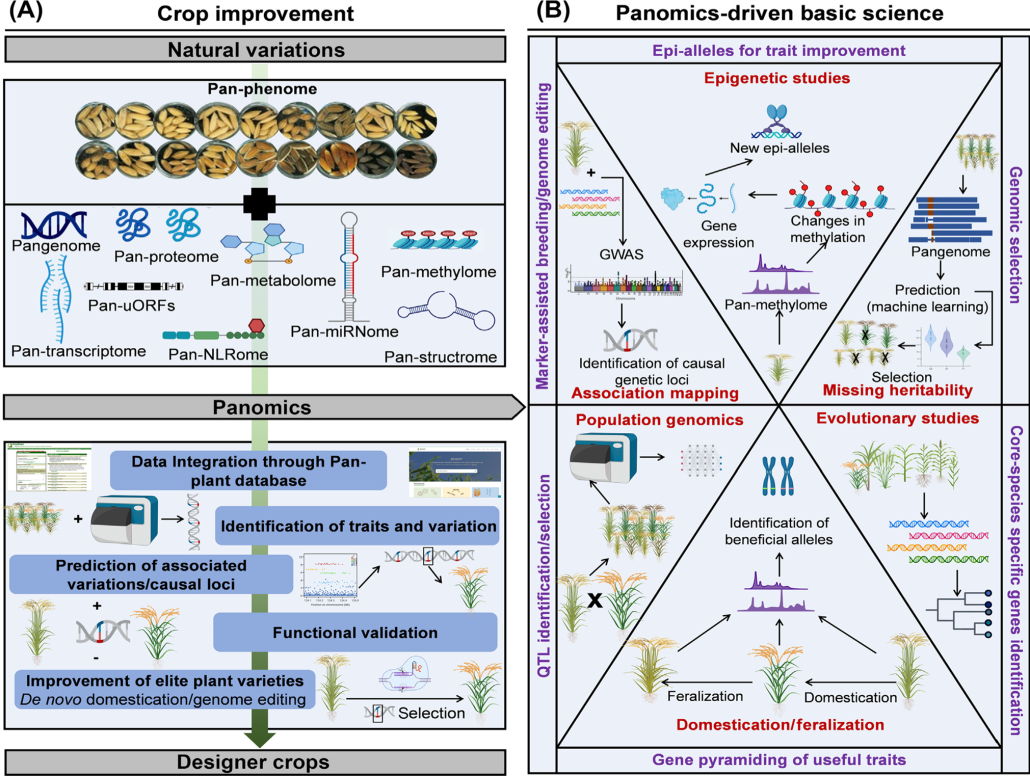
Review: The era of panomics-driven gene discovery in plants
Plant Science Research WeeklyPanomics, an approach integrating multiple ‘omics’ datasets such as genomics, transcriptomics, metabolomics, and phenomics, has seen rapid advancement in recent years due to technological improvements, particularly in genomics. This review focuses on the recent developments in panomics-driven gene…
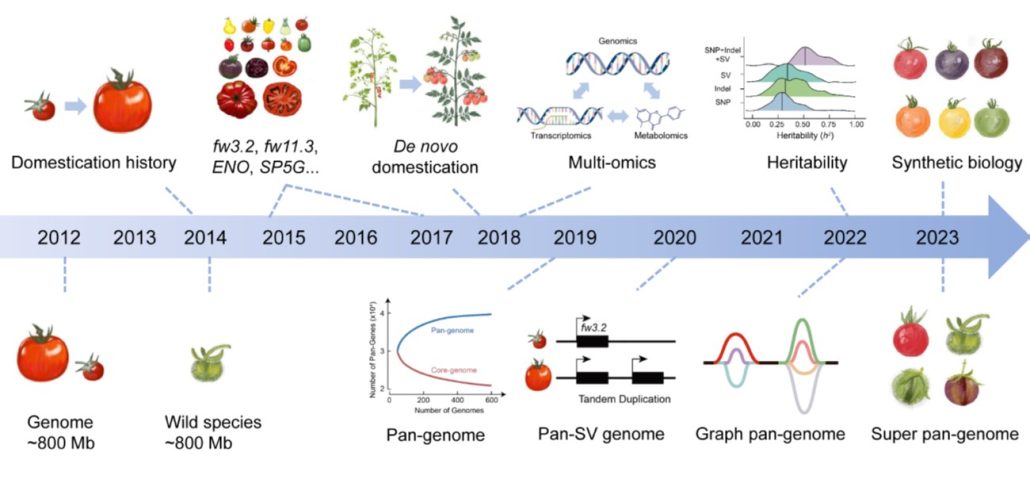
Review: The genomic route to tomato breeding: Past, present, and future
Plant Science Research WeeklyWidely and abundantly eaten tomatoes (Solanum lycopersicum) are delicious and nutritious, but the genetic diversity of cultivated tomatoes is quite narrow. In this review, Wang et al. give an overview of efforts to increase diversity, through introduction of genes from wild relatives and other approaches.…
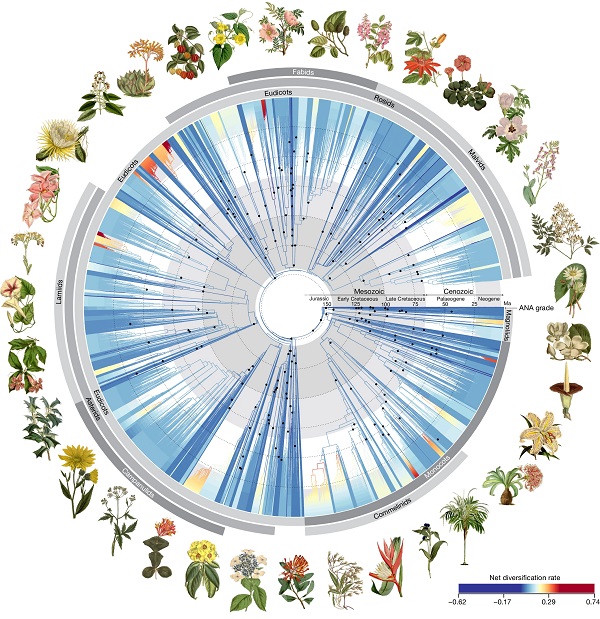
Phylogenomic insights into angiosperm evolution
Plant Science Research WeeklyLow-resolution data can provide broad strokes but miss the details that come from greater information density. When striving to understand the multimillion-year evolutionary history of the angiosperms, more data certainly helps. Here, by focusing on a subset of 353 genes, Zuntini and Carruthers et al.…
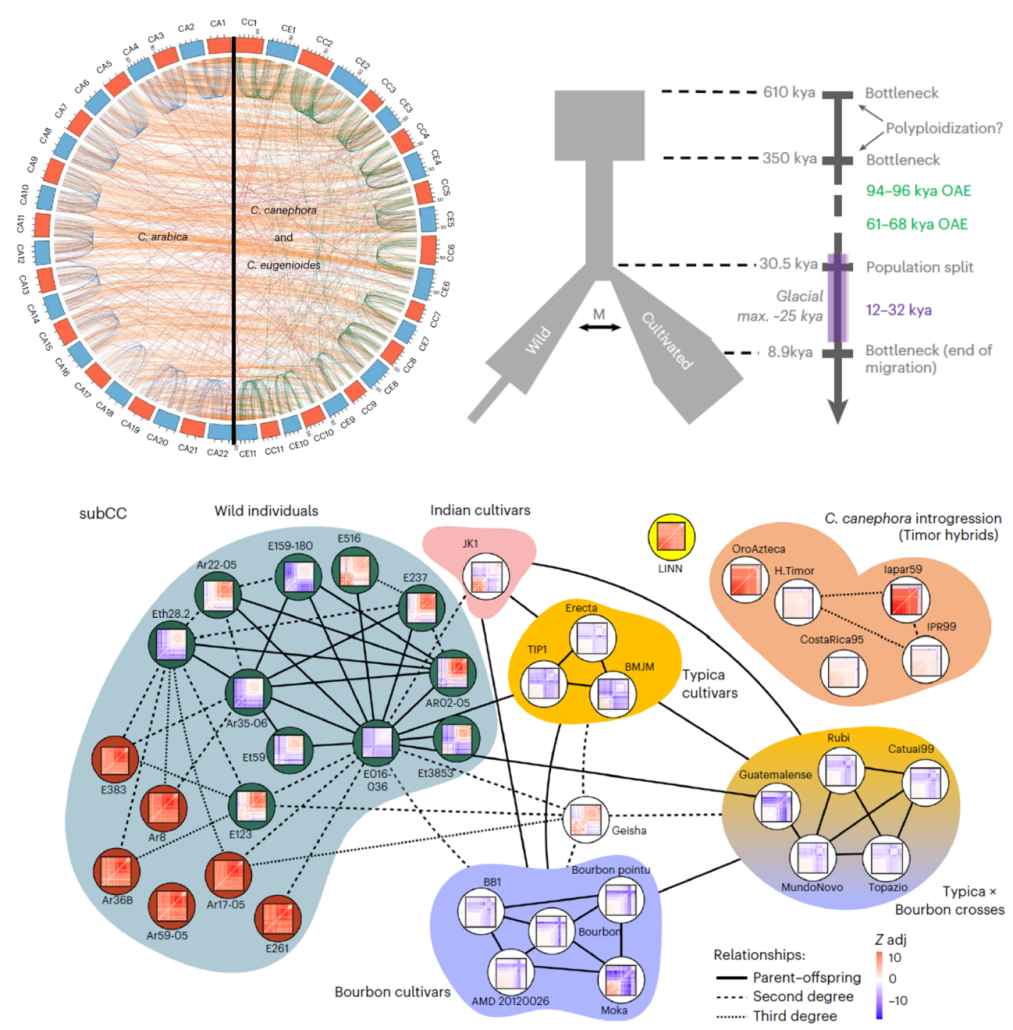
Genomics from bean to cup: New insights into the history of Arabica coffee diversification
Plant Science Research WeeklyAs one of the most traded commodities in the world, coffee has cultural and economic impact that spans continents. The main source of coffee beans, Coffea arabica (Arabica), is a polyploid species that resulted from the hybridization between diploid C. canephora (Robusta) and C. eugenioides (Eugenioides).…

