What We’re Reading: January 5th
Guest Editor: Alecia Biel
 Alecia is a graduate student at The Ohio State University in the US and has been a Plantae Fellow since September 2017. Her research focuses on elucidating hormone signaling pathways and the role of the nucleus during this process, particularly throughout plant abiotic stress responses in guard cells. She is very concerned with how science is perceived by the government and the public and strives to become more involved with science communication and acceptance.
Alecia is a graduate student at The Ohio State University in the US and has been a Plantae Fellow since September 2017. Her research focuses on elucidating hormone signaling pathways and the role of the nucleus during this process, particularly throughout plant abiotic stress responses in guard cells. She is very concerned with how science is perceived by the government and the public and strives to become more involved with science communication and acceptance.
Commentary: Is it ordered correctly? Validating genome assemblies by optical mapping
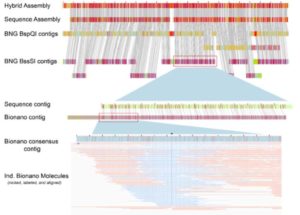 One of the hardest parts of any sequencing project is putting the pieces together. Physical sequence alignments can be cross checked against genetic linkage maps when they are available, but what about for species without genetic linkage data? Udall and Dawe describe the use of optical mapping, using Bionano Genomics or Nabsys technologies, as an additional tool for sequence alignments. These methods use modified restriction enzymes for single-strand nick-based labelling to generate DNA maps. Labeled DNA is passed through nanochannels to generate a map, and the overlapping maps are assembled into an alignment that can be used to validate the physical sequence alignment. In this commentary, the authors describe approaches and limitations to optical mapping for genome assembly. (Summary by Mary Williams) Plant Cell. 10.1105/tpc.17.00514.
One of the hardest parts of any sequencing project is putting the pieces together. Physical sequence alignments can be cross checked against genetic linkage maps when they are available, but what about for species without genetic linkage data? Udall and Dawe describe the use of optical mapping, using Bionano Genomics or Nabsys technologies, as an additional tool for sequence alignments. These methods use modified restriction enzymes for single-strand nick-based labelling to generate DNA maps. Labeled DNA is passed through nanochannels to generate a map, and the overlapping maps are assembled into an alignment that can be used to validate the physical sequence alignment. In this commentary, the authors describe approaches and limitations to optical mapping for genome assembly. (Summary by Mary Williams) Plant Cell. 10.1105/tpc.17.00514.
Evolution of transposon-encoded anti-silencing factors in Arabidopsis ($)
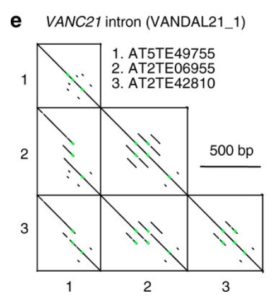 Transposable elements (TEs) are a major component of eukaryotic genomes. Their activity is silenced by epigenetic mechanisms such as chromatin modifications and DNA methylation in order to avoid deleterious effects on host genome stability. Nevertheless, how TEs overcome silencing by the host and propagate is poorly understood. In this study, the authors describe the activity of sequence-specific anti-silencing proteins encoded by Arabidopsis TE families. They show that the anti-silencing VANC21 protein, encoded by members of the TE family VANDAL21, is sufficient for triggering sequence-specific demethylation of VANDAL21 copies. VANC21 binds to specific nine-bases motifs derived from tandem repeats, localized in the non-coding region of VANDAL21. Interestingly, they also identified proteins related to VANC encoded by other TE families, such as the protein VANC6 encoded by VANDAL6 family. VANC6 shows the same behavior as VANC21 and analysis of their target sequences suggests similar evolutionary dynamics. Altogether, this study describes the evolution of TE-specific anti-silencing mechanisms through the behavior of VANC proteins. Such a system could potentially be harnessed to engineer epigenetic states at specific loci, notably in the perspective of genetically modified crops (Summary by Matthias Benoit) Nature Comm. 10.1038/s41467-017-02150-7.
Transposable elements (TEs) are a major component of eukaryotic genomes. Their activity is silenced by epigenetic mechanisms such as chromatin modifications and DNA methylation in order to avoid deleterious effects on host genome stability. Nevertheless, how TEs overcome silencing by the host and propagate is poorly understood. In this study, the authors describe the activity of sequence-specific anti-silencing proteins encoded by Arabidopsis TE families. They show that the anti-silencing VANC21 protein, encoded by members of the TE family VANDAL21, is sufficient for triggering sequence-specific demethylation of VANDAL21 copies. VANC21 binds to specific nine-bases motifs derived from tandem repeats, localized in the non-coding region of VANDAL21. Interestingly, they also identified proteins related to VANC encoded by other TE families, such as the protein VANC6 encoded by VANDAL6 family. VANC6 shows the same behavior as VANC21 and analysis of their target sequences suggests similar evolutionary dynamics. Altogether, this study describes the evolution of TE-specific anti-silencing mechanisms through the behavior of VANC proteins. Such a system could potentially be harnessed to engineer epigenetic states at specific loci, notably in the perspective of genetically modified crops (Summary by Matthias Benoit) Nature Comm. 10.1038/s41467-017-02150-7.
Ash leaf metabolomes reveal differences between trees tolerant and susceptible to ash dieback disease
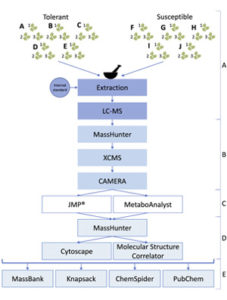 Over the last two decades, Ash dieback (ADB) has been sweeping through Europe killing or damaging a large proportion of European common ash trees (Fraxinus excelsior). ADB results from infection by wind borne spores of the fungus Hymenoscyphus fraxineus. As ADB spread and the scientific research effort increased, it became clear that some ash trees showed tolerance to the fungus and suffered minimal damage. Using transcriptomics/genomics to generate markers for breeding programs is challenging given the high level of genetic heterogeneity in the European ash population, and the fact that resistance to ADB appears to be quantitative. As such, Sambles et al. conducted untargeted QToF mass spectrometry to compare the leaf metabolomes of five tolerant trees with five susceptible trees in a fully-replicated study. The data was used to identify features that were able to discriminate between the tolerant and susceptible samples. For example, iridoid glycosides, a family of terpenoid derivatives with anti-herbivory activity, were shown to be less abundant in leaves from tolerant trees. The data from this study is openly accessible through the Metabolights database for further analysis by the research community (Summary by Mike Page) Sci. Data. 10.1038/sdata.2017.190.
Over the last two decades, Ash dieback (ADB) has been sweeping through Europe killing or damaging a large proportion of European common ash trees (Fraxinus excelsior). ADB results from infection by wind borne spores of the fungus Hymenoscyphus fraxineus. As ADB spread and the scientific research effort increased, it became clear that some ash trees showed tolerance to the fungus and suffered minimal damage. Using transcriptomics/genomics to generate markers for breeding programs is challenging given the high level of genetic heterogeneity in the European ash population, and the fact that resistance to ADB appears to be quantitative. As such, Sambles et al. conducted untargeted QToF mass spectrometry to compare the leaf metabolomes of five tolerant trees with five susceptible trees in a fully-replicated study. The data was used to identify features that were able to discriminate between the tolerant and susceptible samples. For example, iridoid glycosides, a family of terpenoid derivatives with anti-herbivory activity, were shown to be less abundant in leaves from tolerant trees. The data from this study is openly accessible through the Metabolights database for further analysis by the research community (Summary by Mike Page) Sci. Data. 10.1038/sdata.2017.190.
An oomycete plant pathogen reprograms host pre-mRNA splicing to subvert immunity
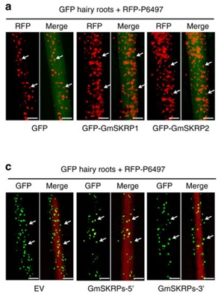 Phytopthora sojae poses a serious threat to soybean production world-wide. This oomycete pathogen has a wide arsenal of effector proteins, some of which have been functionally characterized for their virulence role. Huang et al. characterized and demonstrated the functional role of an avirulence effector PsAvr3c in reprogramming the host RNA-splicing process. PsAvr3c is an essential virulence factor that has a nuclear localization signal and shows interaction with soybean GmSKRP1/2 proteins, the latter associating with plant spliceosome components. PsAvr3c stabilizes the GmSKRP proteins and, being negative regulators of plant immunity, their ectopic expression promotes pathogen colonization. RNA-seq analysis on GmSKRP1 and PsAvr3c overexpression lines indicates a significant alternation in splicing pre-mRNAs, including defense-related genes. Splicing ratios of selected genes show a good co-relation during wild-type and PsAvr3c effector mutant during soybean infections, confirming splicing events to be mediated by PsAvr3c. Hence, this work has shown elucidation of oomycete effectors targeting host pre-mRNA splicing to promote disease (Summary by Amey Redkar) Nat. Comms. 10.1038/s41467-017-02233-5.
Phytopthora sojae poses a serious threat to soybean production world-wide. This oomycete pathogen has a wide arsenal of effector proteins, some of which have been functionally characterized for their virulence role. Huang et al. characterized and demonstrated the functional role of an avirulence effector PsAvr3c in reprogramming the host RNA-splicing process. PsAvr3c is an essential virulence factor that has a nuclear localization signal and shows interaction with soybean GmSKRP1/2 proteins, the latter associating with plant spliceosome components. PsAvr3c stabilizes the GmSKRP proteins and, being negative regulators of plant immunity, their ectopic expression promotes pathogen colonization. RNA-seq analysis on GmSKRP1 and PsAvr3c overexpression lines indicates a significant alternation in splicing pre-mRNAs, including defense-related genes. Splicing ratios of selected genes show a good co-relation during wild-type and PsAvr3c effector mutant during soybean infections, confirming splicing events to be mediated by PsAvr3c. Hence, this work has shown elucidation of oomycete effectors targeting host pre-mRNA splicing to promote disease (Summary by Amey Redkar) Nat. Comms. 10.1038/s41467-017-02233-5.
Oh, the places they’ll go! A survey of phytopathogen effectors and their host targets ($)
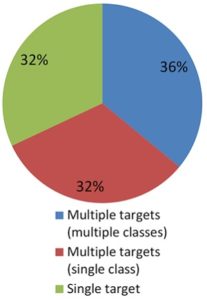 All phytopathogens encode for a toolbox of secreted proteins called ‘effectors’ that promote disease formation in the best possible way. Effectors are either acting in the apoplastic space or are translocated to the host cell to target diverse processes and modulate the host using enzymatic activities. Khan et al. have surveyed the effector literature from numerous phytopathogens and have curated an overview of the locations of effector targets and their molecular and biological functions in terms of similarities and specificities in relation to the pathogen lifestyles. This survey shows that phytopathogens tend to use different immune-suppression strategies, exhibiting diverse properties of effector functions, particularly in targeting plant immunity. This mechanism requires either hijacking multiple components in the host cell or specializing in targeting crucial pressure-points of plant immunity. Hence this work has nicely shed light on the primary role of effectors and additionally points towards interesting specificities, depending on the virulence strategy of the pathogens (Summary by Amey Redkar) Plant Journal. 10.1111/tpj.13780.
All phytopathogens encode for a toolbox of secreted proteins called ‘effectors’ that promote disease formation in the best possible way. Effectors are either acting in the apoplastic space or are translocated to the host cell to target diverse processes and modulate the host using enzymatic activities. Khan et al. have surveyed the effector literature from numerous phytopathogens and have curated an overview of the locations of effector targets and their molecular and biological functions in terms of similarities and specificities in relation to the pathogen lifestyles. This survey shows that phytopathogens tend to use different immune-suppression strategies, exhibiting diverse properties of effector functions, particularly in targeting plant immunity. This mechanism requires either hijacking multiple components in the host cell or specializing in targeting crucial pressure-points of plant immunity. Hence this work has nicely shed light on the primary role of effectors and additionally points towards interesting specificities, depending on the virulence strategy of the pathogens (Summary by Amey Redkar) Plant Journal. 10.1111/tpj.13780.
Disruption of actin filaments in Zea mays by bisphenol A depends on their crosstalk with microtubules
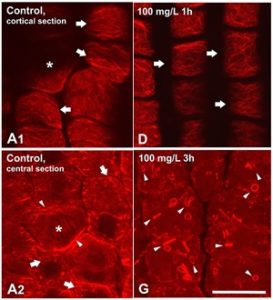 Bisphenol A (BPA) is known for its negative impact on mammalian cell lines and, recently, as an emerging environmental pollutant. BPA, readily taken up and metabolized by plants, results in many growth and developmental defects, from disrupted mitotic microtubule (MT) arrays to altered calcium fluctuations and actin disorganization during pollen tube growth. Stavropoulou et al. found that BPA treatment of Zea mays seedlings decreased germination and root growth, disrupted root MT organization, impaired preprophase band formation, and cells displayed defective cytokinesis. BPA-treated root meristematic protodermal cells exhibited abnormal actin filaments, which were in thick, ring/rod-like confirmations; similarly seen in rhizodermal cells within the elongation zone. These data show that BPA influences the cytoskeleton in dividing plant cells. It is important to understand the environmental impact of BPA as levels are increasing within the water and soil, and this may lead to adverse developmental consequences for many crop species (Summary by Alecia Biel) Chemosphere. 10.1016/J.chemosphere.2017.12.099.
Bisphenol A (BPA) is known for its negative impact on mammalian cell lines and, recently, as an emerging environmental pollutant. BPA, readily taken up and metabolized by plants, results in many growth and developmental defects, from disrupted mitotic microtubule (MT) arrays to altered calcium fluctuations and actin disorganization during pollen tube growth. Stavropoulou et al. found that BPA treatment of Zea mays seedlings decreased germination and root growth, disrupted root MT organization, impaired preprophase band formation, and cells displayed defective cytokinesis. BPA-treated root meristematic protodermal cells exhibited abnormal actin filaments, which were in thick, ring/rod-like confirmations; similarly seen in rhizodermal cells within the elongation zone. These data show that BPA influences the cytoskeleton in dividing plant cells. It is important to understand the environmental impact of BPA as levels are increasing within the water and soil, and this may lead to adverse developmental consequences for many crop species (Summary by Alecia Biel) Chemosphere. 10.1016/J.chemosphere.2017.12.099.
Arabidopsis thaliana plants lacking the ARP2/3 complex show defects in cell wall assembly and auxin distribution
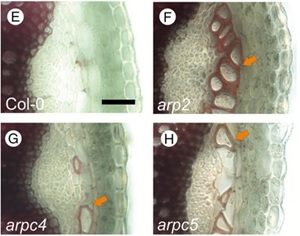 The plant cytoskeleton determines cell shape and integrity by delivering cellulose microfibrils and other cell wall components to the plasma membrane and cell wall. Auxin is involved in establishing the polarity of cell expansion and auxin distribution is partly regulated by actin. Sahi et al. examine the role of ARP2/3 in connecting F-actin dynamics and auxin distribution during cell wall synthesis. Actin nucleation involves the ARP2/3 protein complex and arp2/3 mutants in epidermal cotyledon cells resulted in less lobed cells (increased cell circularity) and cell-cell adhesion defects between epidermal-epidermal and epidermal-guard cells. The arp2/3 mutants also displayed decreased cell wall thickness, altered cellulose content, and reduced auxin transport. These data indicate that ARP2/3 plays a role in controlling cell wall synthesis and auxin transport (Summary by Alecia Biel) Annals of Botany. 10.1093/aob/mcx178.
The plant cytoskeleton determines cell shape and integrity by delivering cellulose microfibrils and other cell wall components to the plasma membrane and cell wall. Auxin is involved in establishing the polarity of cell expansion and auxin distribution is partly regulated by actin. Sahi et al. examine the role of ARP2/3 in connecting F-actin dynamics and auxin distribution during cell wall synthesis. Actin nucleation involves the ARP2/3 protein complex and arp2/3 mutants in epidermal cotyledon cells resulted in less lobed cells (increased cell circularity) and cell-cell adhesion defects between epidermal-epidermal and epidermal-guard cells. The arp2/3 mutants also displayed decreased cell wall thickness, altered cellulose content, and reduced auxin transport. These data indicate that ARP2/3 plays a role in controlling cell wall synthesis and auxin transport (Summary by Alecia Biel) Annals of Botany. 10.1093/aob/mcx178.
KNS4/UPEX1: A Type II Arabinogalactan β-(1,3)-Galactosyltransferase Required for Pollen Exine Development
P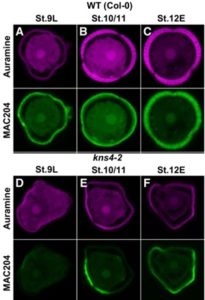 ollen is an essential component in angiosperm reproduction. Pollen grains are surrounded by a highly resistant wall called exine which enables survival of the male gametes in adverse environmental conditions. Suzuki et al. examined the contribution of an Arabinogalactan β-(1,3)-Galactosyltransferase in proper exine development through characterization of the KNS4 gene. The kaonashi mutant (kns4) was initially characterized by abnormal primexine deposition and a lack of proper exine wall patterning, leading to gross morphological defects in the pollen and degradation. This was found to have an impact on fertility as mutant plants yielded shorter siliques and fewer seeds compared to WT. Signal from antibodies against the protein of interest were severely diminished and exhibited altered localization in the mutant. The authors concluded that KNS4 and other arabinogalactan proteins play an important role in angiosperm fertility by promoting pollen exine development (Summary by Andrew Kirkpatrick) Plant Phys. 10.1104/pp.16.01385.
ollen is an essential component in angiosperm reproduction. Pollen grains are surrounded by a highly resistant wall called exine which enables survival of the male gametes in adverse environmental conditions. Suzuki et al. examined the contribution of an Arabinogalactan β-(1,3)-Galactosyltransferase in proper exine development through characterization of the KNS4 gene. The kaonashi mutant (kns4) was initially characterized by abnormal primexine deposition and a lack of proper exine wall patterning, leading to gross morphological defects in the pollen and degradation. This was found to have an impact on fertility as mutant plants yielded shorter siliques and fewer seeds compared to WT. Signal from antibodies against the protein of interest were severely diminished and exhibited altered localization in the mutant. The authors concluded that KNS4 and other arabinogalactan proteins play an important role in angiosperm fertility by promoting pollen exine development (Summary by Andrew Kirkpatrick) Plant Phys. 10.1104/pp.16.01385.
LRX Proteins play a crucial role in pollen grain and pollen tube cell wall development
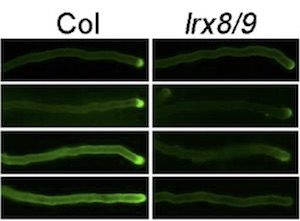 Leucine-rich repeat extensins (LRXs) are proteins involved in cell wall growth and are therefore required for plant growth processes. Fabrice et al. use pollen tube development in Arabidopsis thaliana as a vegetative plant growth model to elucidate the role of LRXs in these cellular processes. All 4 LRX genes expressed in pollen (LRX8, LRX9, LRX10, and LRX11) are redundantly required for pollen tube development. Furthermore, LRX proteins appear to regulate pollen germination, pollen tube growth, composition, and even the ultrastructure of the pollen tube cell wall. Moreover, lrx mutants have altered pollen tube mechanics, showing disrupted vesicle dynamics. This vesicle dynamic phenotype was alleviated in some phenotypes by altering calcium availability. Finally, it appears that the N-terminal leucine-rich repeat domain of LRX proteins are associated with the plasma membrane. This study enhances our understanding of how LRX proteins may be involved in call wall-plasma membrane communication (Summary by Isabel Mendoza) Plant Phys. 10.1104/pp.17.01374.
Leucine-rich repeat extensins (LRXs) are proteins involved in cell wall growth and are therefore required for plant growth processes. Fabrice et al. use pollen tube development in Arabidopsis thaliana as a vegetative plant growth model to elucidate the role of LRXs in these cellular processes. All 4 LRX genes expressed in pollen (LRX8, LRX9, LRX10, and LRX11) are redundantly required for pollen tube development. Furthermore, LRX proteins appear to regulate pollen germination, pollen tube growth, composition, and even the ultrastructure of the pollen tube cell wall. Moreover, lrx mutants have altered pollen tube mechanics, showing disrupted vesicle dynamics. This vesicle dynamic phenotype was alleviated in some phenotypes by altering calcium availability. Finally, it appears that the N-terminal leucine-rich repeat domain of LRX proteins are associated with the plasma membrane. This study enhances our understanding of how LRX proteins may be involved in call wall-plasma membrane communication (Summary by Isabel Mendoza) Plant Phys. 10.1104/pp.17.01374.
Arabidopsis pollen tube integrity and sperm release are regulated by RALF-mediated signaling
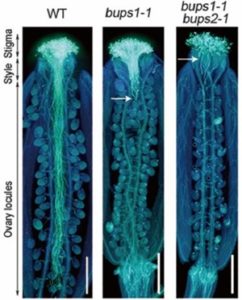 Successful fertilization in plants requires sperm cells (SCs) to be carried along the growing pollen tube (PT) until reaching the female gametophyte where the PT then bursts to release the SCs. One challenge PTs must overcome in order to achieve fertilization is deciding to rupture or not to rupture. Ge et al. identified two receptor-like kinases (RLKs), BUPS1 and BUPS2, expressed at the PT surface. Loss of BUPS1 resulted in shorter siliques and a drastic reduction in seed set, observed to be male-specific. The bups1/2 mutant PTs also ruptured prematurely, most not travelling beyond the style. BUPS1/2 selectively bind to the pollen-expressed peptides RALF4 and RALF19 and ralf4/19 mutants were phenotypically similar to bups1/2 mutants. RALF4/19 also interact with ANX1 and ANX2, two other pollen expressed RLKs. The data suggest a potential heteromer between BUPS1/2 and ANX1/2, which bind RALF4/19 to maintain PT integrity. RALF34, an ovule-derived peptide, was found to induce PT rupture and competes against RALF4/19 for binding to BUPS1 and ANX1. The authors set forth a model in which male-derived ligands bind a PT receptor complex to maintain growth and, upon perception of a female-derived ligand, induces PT rupture (Summary by Alecia Biel) Science. 10.1126/science.aao3642.
Successful fertilization in plants requires sperm cells (SCs) to be carried along the growing pollen tube (PT) until reaching the female gametophyte where the PT then bursts to release the SCs. One challenge PTs must overcome in order to achieve fertilization is deciding to rupture or not to rupture. Ge et al. identified two receptor-like kinases (RLKs), BUPS1 and BUPS2, expressed at the PT surface. Loss of BUPS1 resulted in shorter siliques and a drastic reduction in seed set, observed to be male-specific. The bups1/2 mutant PTs also ruptured prematurely, most not travelling beyond the style. BUPS1/2 selectively bind to the pollen-expressed peptides RALF4 and RALF19 and ralf4/19 mutants were phenotypically similar to bups1/2 mutants. RALF4/19 also interact with ANX1 and ANX2, two other pollen expressed RLKs. The data suggest a potential heteromer between BUPS1/2 and ANX1/2, which bind RALF4/19 to maintain PT integrity. RALF34, an ovule-derived peptide, was found to induce PT rupture and competes against RALF4/19 for binding to BUPS1 and ANX1. The authors set forth a model in which male-derived ligands bind a PT receptor complex to maintain growth and, upon perception of a female-derived ligand, induces PT rupture (Summary by Alecia Biel) Science. 10.1126/science.aao3642.