The rhythm of the light: how light and the clock drive cycling of transcript levels in barley
Circadian rhythms are ubiquitous among living things. A core set of so-called clock genes and their products assemble an oscillatory network that provides a rhythm to anticipate dawn and dusk (Hsu and Harmer, 2014). The circadian clock regulates the day/night rhythms of plants and light itself has many ways of feeding information into the functioning of the clock (Oakenfull and Davis, 2017). If plants are transferred from normal day/night cycles into continuous light these oscillations continue for a while, but the amplitude dampens out and the oscillations disappear after a few days.
Circadian rhythms affect almost all aspects of plant growth, from photosynthesis and flowering to responses to biotic and abiotic stress (Greenham and Mcclung, 2015). So perhaps not surprisingly, in crop plants domestication often selected for mutants in circadian genes. Clock gene mutations in various crop species alter flowering time and growth cycles (Bendix et al., 2015), and these alterations in many species help adapt plants native to equatorial regions to the long day photoperiods and short growing seasons of more northern regions, such as in the case of tomato (Solanum lycopersicum; Müller et al., 2015) and barley (Hordeum vulgare; Faure et al., 2012; Campoli et al., 2013).
 In this issue of Plant Physiology Müller et al. show how transcriptomics, combined with the modelling of a gene regulatory network can provide a flying start into unraveling the inner workings of the circadian clock in barley. On top of that they are able to show the importance of light in driving circadian rhythms, compared to role of the clock gene network in doing so.
In this issue of Plant Physiology Müller et al. show how transcriptomics, combined with the modelling of a gene regulatory network can provide a flying start into unraveling the inner workings of the circadian clock in barley. On top of that they are able to show the importance of light in driving circadian rhythms, compared to role of the clock gene network in doing so.
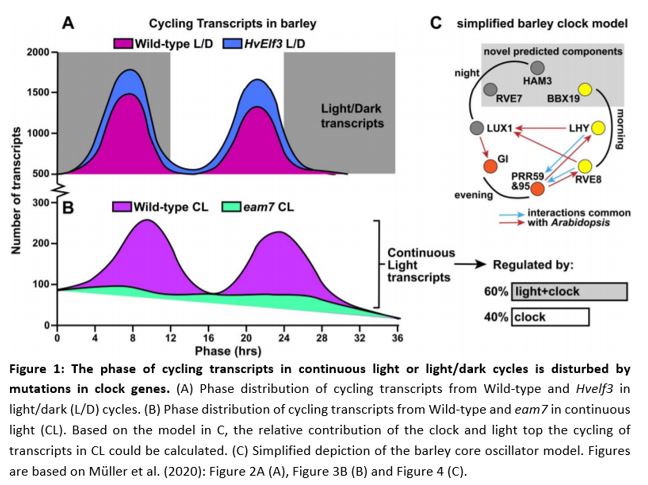
As a base dataset the authors performed an experiment to map cycling genes in day/night and continuous light conditions. RNA sequencing showed that, in wild type cultivar Bowman, the expression of 84% of genes followed a rhythmic day/night cycle (Figure 1A, wild-type). During 36 hours of continuous light, 20% of all gene expression still showed cycling behavior (Figure 1B, wild-type). Plants mutated in HvELF3 and HvLUX1 (orthologs well-known clock genes from Arabidopsis thaliana, EARLY FLOWERING3 and LUX ARRYTHMO) did not exhibit any cycling in gene expression in continuous light. The cycling of these circadian regulated genes tends to follow a bimodal pattern: transcripts increase at the end of night, and again at the end of day. The aforementioned 20% of genes that cycled in continuous light in Bowman showed this bimodal behavior. However, in early maturity 7 (eam7) mutants, this bimodal pattern was lost completely (Figure 1B). Interestingly, Hvelf3 in normal day/night cycles the amount of cycling transcripts had a higher peak at dawn and a higher peak at dusk compared to Bowman, showing a pronounced bimodal pattern (Figure 1A). These data showed that the core clock genes in barley influence the bimodal phase of cycling genes and the light/dark cues determine for the most part if genes cycle or not.
To be able to generate new hypotheses and novel clock genes from their RNA sequencing experiment, the authors created a computational model that showed the regulatory relationships of the barley clock without a-priori knowledge of the network (Mombaerts et al., 2019). As an input for the model, they used genes that showed cycling behavior in continuous light and were common between Bowman and eam7. To avoid computational limitations, they filtered their gene set by searching for Arabidopsis orthologs that were classified by gene ontology as being ‘circadian’ with a high signal/noise ratio in the circadian cycle. In an ideal world this network would have been centered around HvLUX1 and HvELF3, since these genes are indispensable for continuous light cycling transcripts; however only HvLUX1 could be used, because the signal/noise ratio for HvELF3 transcripts was too low.
By aligning the genetic and expression data from Arabidopsis with that of barley, they generated a model that predicts the degree of commonality to the Arabidopsis clock. Some components are missing, but the model nonetheless predicts a number of novel genes involved in oscillatory regulation. HvBBX19, HvRVE7 and HvHAM3 were, as of yet, unknown as clock components; however, these genes can now be tested to investigate their function in governing the cycling rhythms of genes in barley (Figure 1C).
With the help of the authors’ models, it was also possible to investigate the proportion of transcripts cycling in continuous-light that were regulated primarily by the circadian clock compared with the proportion regulated primarily by the light in the normal day/night cycle. It turns out that about 40% reveal a phase dominated by the circadian clock and the remaining 60% of transcripts are under the control of light signaling pathways or co-regulated by light signaling and clock (Figure 1B). Basically, this means that the clock helps transcripts cycle, but the light sets the time!
In conclusion, this work by Müller et al. gives plant scientists new tools to investigate circadian rhythms of barley. This is also important for plant breeding and agronomy since many cultivars of other (grain) crops like barley have mutations in genes considered as part of the core clock (Bendix et al., 2015). Therefore, knowledge of the clock can help adapt species to new growing environments.
Kasper van Gelderen; [email protected]
Department of Biology, Utrecht University, the Netherlands
The author wishes to declare no competing interests.
Literature Cited
Bendix C, Marshall CM, Harmon FG (2015) Circadian Clock Genes Universally Control Key Agricultural Traits. Mol Plant 8: 1135–1152
Campoli C, Pankin A, Drosse B, Casao CM, Davis SJ, Von Korff M (2013) HvLUX1 is a candidate gene underlying the early maturity 10 locus in barley: Phylogeny, diversity, and interactions with the circadian clock and photoperiodic pathways. New Phytol 199: 1045–1059
Faure S, Turner AS, Gruszka D, Christodoulou V, Davis SJ, Von Korff M, Laurie DA (2012) Mutation at the circadian clock gene EARLY MATURITY 8 adapts domesticated barley (Hordeum vulgare) to short growing seasons. Proc Natl Acad Sci U S A 109: 8328–8333
Greenham K, Mcclung CR (2015) Integrating circadian dynamics with physiological processes in plants. Nat Publ Gr 16: 598–610
Hsu PY, Harmer SL (2014) Wheels within wheels: The plant circadian system. Trends Plant Sci 19: 240–249
L.M. Müller, Mombaerts L, A. Pankin, Davis SJ, Webb AAR, Goncalves J, Korff M von (2020) Differential effects of day-night cues and the circadian clock on the barley transcriptome. Plant Physiol. https://doi.org/10.1104/pp.19.01411
Mombaerts L, Carignano A, Robertson FR, Hearn TJ, Junyang J, Hayden D, Rutterford Z, Hotta CT, Hubbard KE, Maria MRC, et al (2019) Dynamical differential expression (DyDE) reveals the period control mechanisms of the Arabidopsis circadian oscillator. PLoS Comput Biol 15: 1–26
Müller NA, Wijnen CL, Srinivasan A, Ryngajllo M, Ofner I, Lin T, Ranjan A, West D, Maloof JN, Sinha NR, et al (2015) Domestication selected for deceleration of the circadian clock in cultivated tomato. Nat Genet 48: 89–93
Oakenfull RJ, Davis SJ (2017) Shining a light on the Arabidopsis circadian clock. Plant Cell Environ 2571–2585