Predicting Gene Regulatory Networks
Zhou et al. develop and test potential gene regulatory networks in maize. Plant Cell https://doi.org/10.1105/tpc.20.00080
By Peng Zhou and Nathan Springer
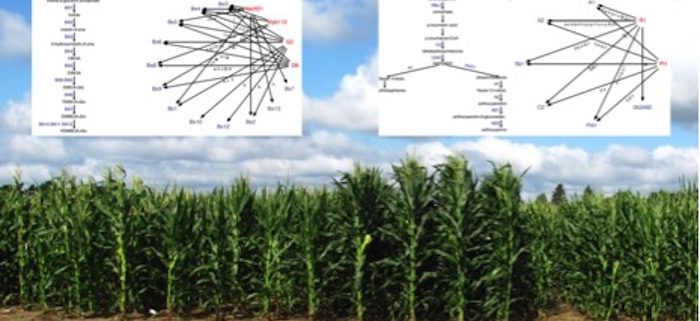
Background: Transcription factors (TFs) play critical roles in regulating the expression of genes. A single TF can control the expression of many genes, including instances in which a TF regulates many genes with roles in the same metabolic pathway. One key step in understanding transcriptional regulation is to define the gene regulatory networks (GRNs) that describe the interactions between TFs and target genes. One approach to predicting GRNs is to use the co-expression patterns of TFs and other genes to predict potential regulatory interactions. Many groups have documented genome-wide expression profiles for various tissues and genotypes of maize, and these were used to develop a set of GRN predictions.
Question: We wanted to compare the computationally inferred TF-target interactions with experimentally derived validations: What are the characteristics of computational predictions? Do they contain useful biological information, especially about linking TFs to the regulation of multiple genes in a pathway? We also wanted to explore the difference between networks built using different types of expression datasets. Finally, does the information from these GRNs accurately predict natural variation?
Findings: We found that GRNs created from different types of datasets are enriched for biologically relevant interactions, but different networks capture distinct TF-target associations and biological processes. When using natural variation datasets to test the predictions of GRNs, we found that presence/absence changes in expression of a TF, rather than quantitative changes in TF expression, are more likely associated with changes in target gene expression. The integration of our TF-target predictions with pathway annotations and previous results provide support for many TFs and their putative targets spanning a variety of plant metabolic pathways.
Next steps: Our findings provide potential targets for breeding or biotechnological applications. Given the prediction of specific TFs that may play roles in regulating multiple target genes from a pathway or physiological process, we can predict key TFs that may be good targets for crop improvement. There is also room for substantial improvement in GRN prediction, including developing models that would incorporate additional types of data and machine-learning methods that may synthesize information across networks and data types.
Peng Zhou, Zhi Li, Erika Magnusson, Fabio A. Gomez Cano, Peter A. Crisp, Jaclyn M. Noshay, Erich Grotewold, Candice N. Hirsch, Steven P. Briggs, Nathan M. Springer (2020). Meta Gene Regulatory Networks in Maize Highlight Functionally Relevant Regulatory Interactions. Plant Cell: DOI: https://doi.org/10.1105/tpc.20.00080