The Multifaceted Roles of Autophagy in Fixed-carbon Starvation
McLoughlin et al. uncover the critical roles of autophagy in recycling amino acids and nitrogen-rich nucleotides, adjusting respiratory substrates, and the retention of assimilated nitrogen in maize during fixed-carbon starvation. The Plant Cell (2020) https://doi.org/10.1105/tpc.20.00226
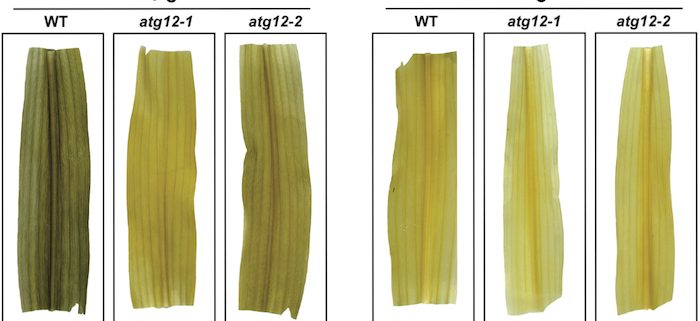
Background: Autophagy is a central recycling system that helps remove unnecessary or damaged intracellular material, including organelles, protein complexes, lipid bodies, and protein aggregates, through the capture and vesicular transport of selected cargo to the vacuole for degradation. This clearance is vital not only for minimizing cytotoxicity, but also for the efficient reuse of nutrients needed for new growth. Accordingly, plants lacking autophagy have increased levels of proteins and various lipid metabolites, are hypersensitive to nutrient starvation and less fecund, and senesce prematurely. Here, we focus on how autophagy helps maize respond to and survive strong fixed-carbon deficits induced by darkness using an integrated omics approach combining deep metabolomic, proteomic, transcriptomic, and ionomic datasets obtained from a strong autophagy mutant.
Question: How does maize cope with fixed-carbon limitation normally and in the absence of autophagy? Does autophagy of proteins under these conditions occur in bulk, or is it driven by selective mechanisms? Can we identify regulators and compensatory pathways that can be exploited to make crops more resilient during nutritional stress?
Findings: Fixed-carbon starvation amplifies autophagic turnover in maize leaves to compensate for this deficiency, which in turn initiates several compensatory responses when this recycling is blocked. Included is an acceleration of complex carbohydrate, lipid, and nucleotide catabolism diverted to maintain fatty acid b-oxidation and mitochondrial respiration, combined with mechanisms to capture the released nitrogen in asparagine, glutamine, and the ureides allantoin and allantoic acid. The mutant plants also hyperaccumulate a wide variety of proteins that are presumed targets of autophagy, including those contained within peroxisomes and mitochondria and the ribosome and proteasome complexes, and dramatically increase the levels of anti-oxidants such a glutathione to protect against oxidative damage. By combining the metabolomic, proteomic and transcriptomic datasets, key nutritional regulatory points in maize metabolism emerged. is a negative regulator of spikelet number per panicle. Altering this gene directly influenced panicle patterning. We discovered that OsER1 acts upstream of the OsMKKK10-OsMKK4-OsMPK6 kinase cascade. Surprisingly, the OsER1-OsMKKK10-OsMKK4-OsMPK6 pathway is required for the maintenance of cytokinin homeostasis.
Next steps: Understanding the impacts of autophagy during stress has revealed regulatory priorities, bottlenecks, and alternative pathways used by plants to maintain growth and stress resilience. By manipulating autophagy, it might be possible to improve the productivity and nutrient-use efficiency of crops when grown under suboptimal conditions or with less fertilizer input.
Fionn McLoughlin, Richard S. Marshall, Xinxin Ding, Elizabeth C. Chatt, Liam D. Kirkpatrick, Robert C. Augustine, Faqiang Li, Marisa S. Otegui, and Richard D. Vierstra (2020). Autophagy Plays Prominent Roles in Amino Acid, Nucleotide, and Carbohydrate Metabolism During Fixed-carbon Starvation in Maize. Plant Cell DOI: https://doi.org/10.1105/tpc.20.00226