A Hub of Hubs: The Central Role of ZmABI19 in the Regulatory Network of Maize Grain Filling
Cereal grains accumulate large amounts of proteins and starch and are a major source of dietary calories for humans and animals. In maize (Zea mays), most of the storage compounds are synthesized during the grain-filling period of kernel development. Several key transcription factors (TFs) that regulate grain-filling have been discovered (reviewed in Li and Song, 2020), including a few—Opaque2 (O2), Prolamin-box factor 1 (PBF1), ZmNAC128, ZmNAC130, and Opaque11 (O11)—that show endosperm-specific and coordinated accumulation patterns, suggesting that they are regulated by a common set of upstream regulators.
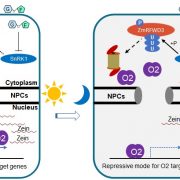
 In this issue of The Plant Cell, first authors Tao Yang, Liangxing Guo and colleagues (Yang et al., 2021) set out to identify some of these upstream regulators by screening for cis-elements in the O2 promoter that are crucial for trans-activating O2. The authors identified two copies of the RY motif, a known DNA-binding site for the plant-specific B3 family of TFs. A series of protein-DNA interaction assays quickly focused on ZmABI19 as the only B3 protein in maize that specifically binds these two RY motifs. An expression analysis using multiple maize tissues/organs showed that ZmABI19 transcripts predominantly accumulate in the kernel.
In this issue of The Plant Cell, first authors Tao Yang, Liangxing Guo and colleagues (Yang et al., 2021) set out to identify some of these upstream regulators by screening for cis-elements in the O2 promoter that are crucial for trans-activating O2. The authors identified two copies of the RY motif, a known DNA-binding site for the plant-specific B3 family of TFs. A series of protein-DNA interaction assays quickly focused on ZmABI19 as the only B3 protein in maize that specifically binds these two RY motifs. An expression analysis using multiple maize tissues/organs showed that ZmABI19 transcripts predominantly accumulate in the kernel.
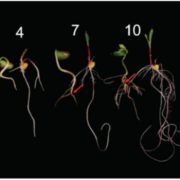
To functionally characterize ZmABI19, the authors created two zmabi19 mutant alleles using clustered regularly interspaced short palindromic repeats (CRISPR) and the CRISPR-Associated nuclease Cas9. Both were loss-of-function alleles and resulted in smaller and flattened kernels relative to wild type. One of the alleles, zmabi19-1, was chosen for further analyses. The mutant kernels were opaque, with a significantly reduced 100-kernel weight and starch/zein/lipid contents. The mRNA and protein levels of O2, Pbf1, and nearly all zein families were significantly down-regulated in the mutant. The authors also showed that ZmABI19 binds an RY motif in the Pbf1 promoter, suggesting that ZmABI19 directly regulates both O2 and Pbf1 and acts as an initiator of grain-filling. Furthermore, the mutant exhibited a slower endosperm development and arrested development of the embryo, causing seed lethality.
To understand the regulatory roles of ZmABI19, the authors then carried out a series of deep transcriptome sequencing (RNA-seq) and chromatin immunoprecipitation followed by deep sequencing (ChIP-seq). RNA-seq of 16-DAP endosperm and embryo from the zmabi19-1 mutant showed that the loss of ZmABI19 function significantly impaired protein and starch biosynthesis. Combined RNA-seq and ChIP-seq analyses of 8-DAP kernels identified 106 putative direct target genes for ZmABI19, including nearly all the known grain-filling regulators mentioned above (see Figure). This result indicates that ZmABI19 is a central regulator of grain filling. Moreover, the authors found that ZmABI19 directly regulates Viviparous1 (encoding a TF regulator of scutellum development and intra-kernel protein re-allocation), SUGARS WILL EVENTUALLY BE EXPORTED TRANSPORTER 4c (SWEET4c, encoding a sugar transporter involved in grain-filling and cell differentiation of the basal endosperm transfer layer), and several auxin signaling pathway genes, suggesting a broad role of ZmABI19 in the regulation of kernel development.
A few B3 family TFs that are evolutionarily related to ZmABI19, namely LEAFY COTYLEDON2 (LEC2), ABSCISIC ACID INSENSITIVE 3 (ABI3), and FUSCA3 (FUS3), had been suggested to be key regulators of seed development in eudicots such as Arabidopsis (Arabidopsis thaliana). Supported by the highlighted work and other studies, the functions of these TFs are to some extent conserved in monocots and eudicots (reviewed in Sreenivasulu and Wobus, 2013). Therefore, further investigation of the related TFs and the gene networks they regulate in maize and other seed crops has the potential to contribute to future improvement of grain yield and quality.
Junpeng Zhan
Donald Danforth Plant Science Center
St. Louis, Missouri
Department of Biology and Institute of Plant and Food Science
Southern University of Science and Technology
Shenzhen, Guangdong, China
jzhan@danforthcenter.org
ORCID ID: 0000-0001-7353-7608
REFERENCES
Li, C., and Song, R. (2020). The regulation of zein biosynthesis in maize endosperm. Theoretical and Applied Genetics 133: 1443-1453.
Sreenivasulu, N., and Wobus, U. (2013). Seed-Development Programs: A Systems Biology-Based Comparison Between Dicots and Monocots. Annual Review of Plant Biology 64: 189-217.
Yang, T., Guo, L., Ji, C., Wang, H., Wang, J., Zheng, X., Xiao, Q., and Wu, Y. (2021). The B3 Domain Transcription Factor ZmABI19 Coordinates Expression of Key Factors Required for Maize Seed Development and Grain Filling. Plant Cell, https://bit.ly/34lY4d0.