Genomes losing balance – what is the consequence?
By Xiaowen Shi and James A. Birchler
Division of Biological Sciences, University of Missouri, Columbia, MO 65211
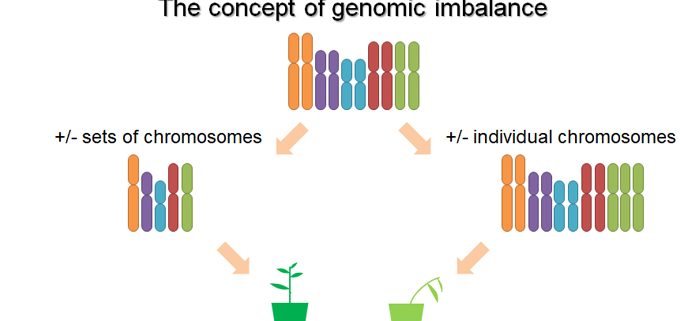
Background: A change in the number of chromosomes can cause problems with growth, development, and function in an organism. One famous example would be Down Syndrome, one of the most common human conditions due to changes in chromosomal dosage. There are two types of abnormality of chromosomal dosage: one is changing the dosage of individual chromosomes or chromosomal segments (aneuploidy); the other is altering the dosage of whole set of chromosomes (polyploidy). Typically, aneuploidy has more phenotypic defects than polyploidy. This phenomenon became known as genomic imbalance and a gene balance hypothesis was proposed to explain this observation. In an aneuploid condition, the quantitative relationship/ratio of components of protein complexes would be disrupted, thus causing negative consequences. However, this relationship is not affected in polyploidy.
Question: Previous studies focused more on increased chromosomal dosage, while our study contains both increased and decreased chromosomal dosage on a larger scale with varied chromosome regions covering over two-thirds of the maize genome. Therefore, we would be able to investigate how genomic imbalance impacts gene expression across all chromosomes and at the individual gene level.
Findings: We examined the impact of addition or subtraction of sixteen different chromosomal segments by quantifying the level of all mRNAs expressed in maize mature leaf tissue. Our study illustrates that there is an overall trend for greater changes in gene expression in aneuploids than polyploids. It was generally accepted in the field that altered expression in aneuploids mainly occurs on the chromosome with varied dosage. Our results suggest that aneuploidy induces changes in gene expression not only on the varied chromosome but also on the chromosomes whose copy number is not altered. Furthermore, in addition to the most common responses to genomic imbalance, we revealed the existence of increased and decreased effects in which expression of many genes are modulated towards the same direction regardless of increased or decreased chromosomal dosage.
Next steps: To investigate how small RNAs, transposable elements and epigenetic marks respond to genomic imbalance would enable us to see the big picture of how genomic imbalance functions. With the development of new sequencing technologies and the emergence of big data, development of computational tools would facilitate our understanding of the interplay of gene regulatory networks under genomic imbalance.
Xiaowen Shi, Hua Yang, Chen Chen, Jie Hou, Katherine M. Hanson, Patrice S. Albert, Tieming Ji, Jianlin Cheng and James A. Birchler. (2021). Genomic imbalance determines positive and negative modulation of gene expression in diploid maize. The Plant Cell, https://doi.org/10.1093/plcell/koab030