Unraveling bHLH transcription factor-mediated maize anther development
By Chong Teng (DDPSC, MO), Guo-Ling Nan (Stanford University, CA), Blake Meyers (DDPSC and University of Missouri–Columbia, MO), and Virginia Walbot (Stanford University, CA)
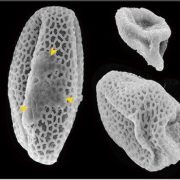
Background: Correct spatiotemporal development of somatic tissues in the anther is necessary for successful fertile pollen production. This development is mediated by many transcription factors acting through complex, multi-layered networks. MS23, MS32, bHLH122, and bHLH51 are sequentially involved in somatic development of the anther lobe based on studies in Arabidopsis, rice, and maize. Both the temporal order in which these four bHLH proteins function and their roles in anther development and 24-nt phased secondary small interfering RNA (phasiRNA) biogenesis are largely unclear. 24-nt phasiRNAs are highly abundant in maize anthers during early meiosis.
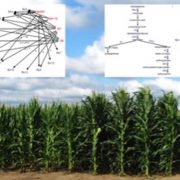
Question: Our goal is to clarify the hierarchy and roles of four maize bHLHs—MS23, MS32, bHLH122, and bHLH51—in tapetal cell differentiation and 24-nt phasiRNA biogenesis.
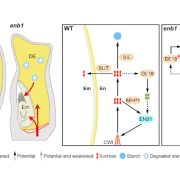
Findings: Functional analysis of knockout mutants in interacting basic-helix-loop-helix genes Ms23, Ms32, bHLH122, and bHLH51 established that male fertility requires all four genes, expressed sequentially in the tapetum. Not only do they regulate each other, but their encoded proteins also form heterodimers that act collaboratively to guide many cellular processes at specific developmental stages. MS23 is confirmed to be the master factor, whose mutation leads to the earliest developmental defect, which is cytologically visible in the tapetum; ms23 mutants exhibit the most drastic alterations in pre-meiotic anther gene expression. Mutations in the four bHLH genes result in mis-regulation of more than 800 downstream transcription factors. The male sterile ms23, ms32, and bhlh122-1 mutants lack 24-nt phasiRNAs and the precursor transcripts from the corresponding 24-PHAS loci. The bhlh51-1 mutant has wild-type levels of both precursors and small RNA products. Both MS23 and MS32 activate Dcl5, which encodes an endoribonuclease that cleaves 24-PHAS precursors, producing 24-nt phasiRNAs. MS23 and MS32 are also required for the transcription of most 24-PHAS loci, with additional and distinctive action by bHLH122 in regulating these loci.
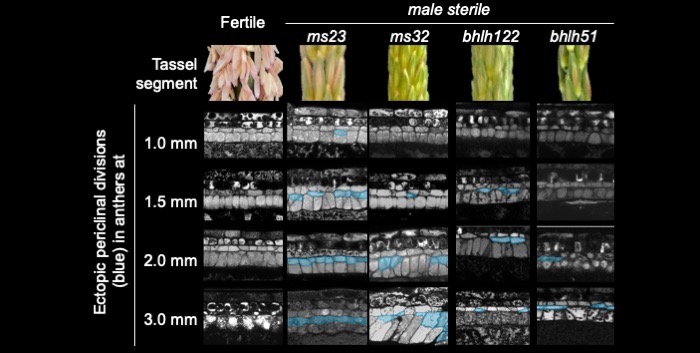
Next steps: We plan to identify the direct targets of each bHLH in order to organize a more detailed transcription factor hierarchy in the context of anther development and 24-nt phasiRNA biogenesis. We would also like to apply this knowledge to improve crop yield.
Guo-Ling Nan, Chong Teng, John Fernandes, Lily O’Connor, Blake C. Meyers, and Virginia Walbot (2022). A cascade of bHLH-regulated pathways program maize anther development. Plant Cell. DOI: 10.1093/plcell/koac007