Tectonic Movements Across the Tomato Genome
Corem et al. demonstrate a switch in DNA methylation patterns across the tomato genome. The Plant Cell (2018). https://doi.org/10.1105/tpc.18.00167.
By Shira Corem, Tzahi Arazi and Nicolas Bouché
Background: Crop genomes are covered by transposons, which are DNA sequences that can jump from one place to another. Transposon mobility must be tightly controlled to avoid deleterious mutations and genome instability. This is especially critical in tomato where transposons are very abundant, not only near the chromosome center, where they are usually concentrated in regions called “heterochromatin,” but also at chromosome arms in the “euchromatin” that contains the genes. One function of cytosine DNA methylation is to stabilize the genome by silencing transposons. In plants, the methylation of euchromatic transposons is mainly related to the constant production of small 20- to 24-nucleotide-long RNA molecules that are driving a specific type of de novo cytosine methylation, restricted to these regions. The vast majority of heterochromatic transposons are kept silent over generations by other mechanisms of cytosine methylation controlled by enzymes like DECREASE IN DNA METHYLATION1 (DDM1).
Question: Our research was aimed at elucidating the way transposons are controlled in a complex genome like tomato, by perturbing DDM1 function. How do plants cope with such harmful mobile elements when the main mechanism controlling their silencing is deficient?
Findings: We utilized CRISPR/Cas9 technology to generate knockout mutants of the two DDM1 genes identified in tomato, and generated a double mutant by crossing them. While each single ddm1 mutant was indistinguishable from wild-type plants, the double mutant displayed pleiotropic developmental phenotypes, associated with severe loss of DNA methylation found in the heterochromatic transposons. Interestingly, euchromatic transposons, controlled by small RNAs and not DDM1, also lost their capacity to be de novo methylated. We discovered that the ddm1 cells start to remethylate transposons of heterochromatin at the expense of euchromatic transposons. When the control of heterochromatin is severely impaired, the silencing mechanism normally controlling euchromatic transposons is partially retargeted towards heterochromatin, thus limiting the consequences of a major transposon awakening.
Next steps: We aim to evaluate of the function of additional genes regulating cytosine methylation in tomato, towards a deeper understanding of epigenetic regulation in a model crop with a complex genome.
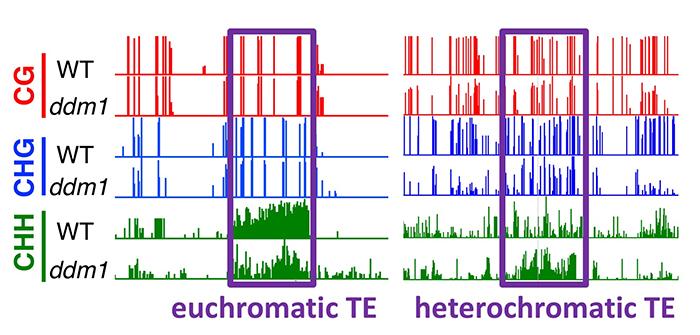
Corem, S., Doron-Faigenboim, A., Jouffroy, O., Maumus, F., Arazi, T., and Bouché, N. (2018). Redistribution of CHH methylation and small interfering RNAs across the genome of tomato ddm1 mutants. Plant Cell. Published June 2018. DOI: https://doi.org/10.1105/tpc.18.00167