Metabolic Signaling Regulates Alternative Splicing during Photomorphogenesis
IN BRIEF by Kathleen L. Farquharson [email protected]
Alternative splicing (AS) regulates gene expression and greatly expands the coding capacity of complex genomes. By regulating which elements of an mRNA transcript are removed or retained, AS produces multiple transcripts from a single gene. Some of these transcripts contain a premature termination codon (PTC) and are targeted for degradation by nonsense-mediated mRNA decay (NMD; Kalyna et al., 2012), whereas others give rise to proteins with various structures and functions. Given that AS has fundamental roles in a wide range of plant growth and development processes (Staiger and Brown, 2013), it is not surprising that over 60% of multiexonic Arabidopsis thaliana genes are regulated by this mechanism (Marquez et al., 2012). Several studies indicate that light conditions dramatically affect AS in plants. Shikata et al. (2014) reported that 6.9% of genes in the genome of etiolated Arabidopsis seedlings exhibit AS changes within 1 h of exposure to red light and demonstrated that these changes occurred in a phytochrome-dependent manner. Petrillo et al. (2014) also reported that light/dark transitions affect the AS of a subset of Arabidopsis genes, but showed that this change was regulated by chloroplast retrograde signaling, which communicates the cell’s energy status to the nucleus.
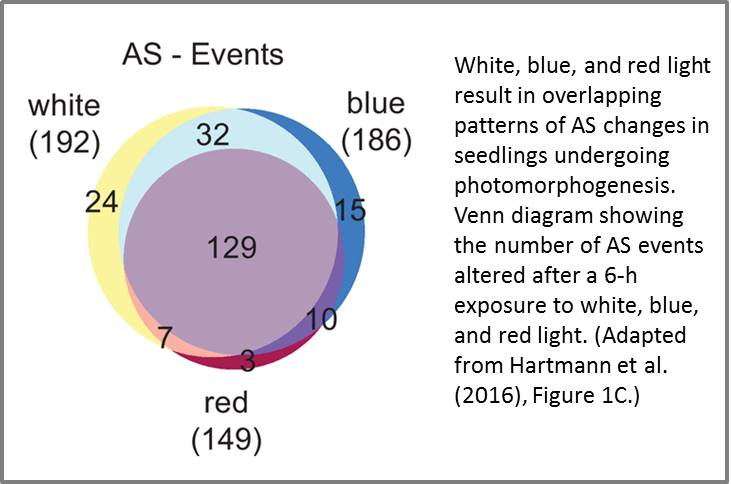
To examine how AS contributes to photomorphogenesis and investigate the mechanism regulating light-modulated AS, Hartmann et al. (2016) compared the transcriptomes of etiolated Arabidopsis seedlings exposed to red, blue, or white light. RNA-sequencing analysis revealed changes in several hundred AS events following a 6-h exposure to at least one of the three light qualities. Although many AS events were altered under all three light conditions, red light triggered fewer AS changes than did blue and white light (see figure). The authors then examined if the changes in AS affected the proportion of transcripts harboring NMD triggering properties. Strikingly, ∼60% of the light-regulated AS events resulted in a reduction of the splicing variant with NMD features and an increase of the putative protein-coding variant. Thus, light exposure appears to boost levels of functional mRNA variants.
Next, the authors conducted a series of experiments to identify the upstream regulators of light-modulated AS. A comparison of the AS profiles of wild-type seedlings and various photoreceptor mutants following illumination with different wavelengths of light suggested that phytochromes and cryptochromes are only minor regulators of white light-modulated AS changes. Intriguingly, etiolated seedlings grown in the presence of sucrose exhibited shifts in AS that resembled those observed following exposure to light. In agreement with the findings of Petrillo et al. (2014), this suggests that metabolic signaling is a major upstream regulator of light-modulated changes in AS. Hartmann et al. further showed that kinase signaling could be an important component of the pathway regulating AS changes in seedlings exposed to light or grown in the presence of sucrose.
Based on these findings, the authors propose that light-modulated AS changes are largely regulated by the plant’s energy status and that photoreceptor signaling is a minor regulator of these changes. This work provides exciting clues as to how plants ramp up the production of key proteins during photomorphogenesis.
REFERENCES
Hartmann, L., Drewe-Boß, P., Wießner, T., Wagner, G., Geue, S., Lee, H.-C., Obermüller, D.M., Kahles, A., Behr, J., Sinz, F.H., Rätsch, G., Wachter, A. (2016). Alternative splicing substantially diversifies the transcriptome during early photomorphogenesis and correlates with the energy availability in Arabidopsis. Plant Cell 28: 2715–2734.
Kalyna, M., et al. (2012). Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 40: 2454–2469.
Marquez, Y., Brown, J.W., Simpson, C., Barta, A., and Kalyna, M. (2012). Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 22: 1184–1195.
Petrillo, E., Herz, M.A., Fuchs, A., Reifer, D., Fuller, J., Yanovsky, M.J., Simpson, C., Brown, J.W., Barta, A., Kalyna, M., and Kornblihtt, A.R. (2014). A chloroplast retrograde signal regulates nuclear alternative splicing. Science 344: 427–430.
Shikata, H., Hanada, K., Ushijima, T., Nakashima, M., Suzuki, Y., and Matsushita, T. (2014). Phytochrome controls alternative splicing to mediate light responses in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 18781–18786.
Staiger, D., and Brown, J.W.S. (2013). Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 25: 3640–3656.





Leave a Reply
Want to join the discussion?Feel free to contribute!