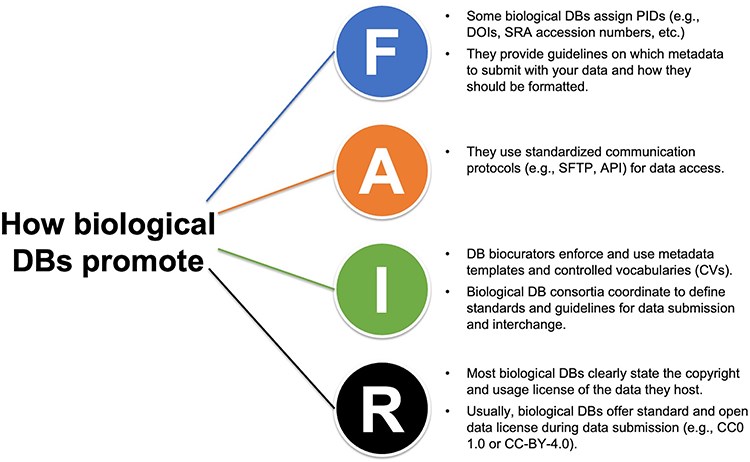
Training scientists to make their data FAIR
Plant Science Research WeeklyI’m sure many of you have experienced frustration when trying to access an intriguing dataset that either doesn’t exist, isn’t open, or is set up in an impossibly unintuitive manner. Part of that problem stems from a lack of training of early-career scientists in how to make their data findable,…
Review: Deep learning approaches to understanding stomatal function
Plant Science Research WeeklySydney Brenner famously said, “Progress in science depends on new techniques, new discoveries and new ideas, probably in that order.” Right now, we’re seeing how advancements and new techniques in artificial intelligence and deep learning are being applied in plant sciences, including in the analysis…
Identifying candidates from genome wide association studies using gene orthologs
Plant Science Research WeeklyGenome wide association studies (GWAS) identify genomic loci associated with a specific trait. However, these loci often contain many genes, so selecting which to investigate further can be tricky. To improve this Whitt, et al. developed a program called FiReMAGE (filtering results of multi-species,…

JGI Plant Gene Atlas: an updateable transcriptome resource to improve functional gene descriptions across the plant kingdom
Plant Science Research WeeklyFunctional genomic studies across plants are incomplete without deciphering the putative gene functions in model as well as non-model plants. With an aim to facilitate gene function identification and cross-species expression analysis, Sreedasyam et al. describe the Joint Genome Institute (JGI) Plant…

Review: Computer models of cell polarity establishment in plants
Plant Science Research WeeklyIt’s pretty obvious that plants are not simply balls of cells. Their shapes and patterns are determined in large part through processes that cause cells to grow and divide asymmetrically, through the establishment of cell polarity. When I set out to read this review I was a bit nervous, expecting to…
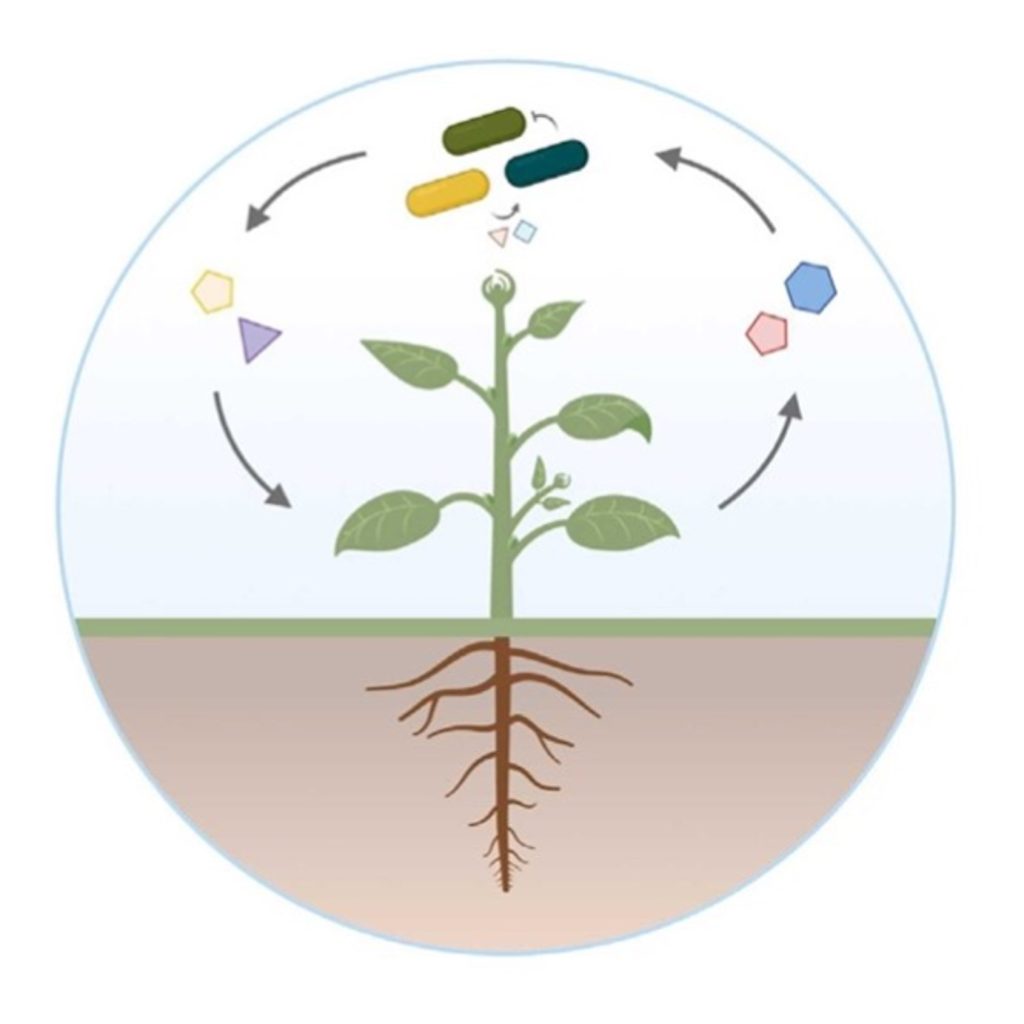
Review: Resolving metabolic interaction mechanisms in plant microbiomes
Plant Science Research WeeklyLife is spurred forward by the power of metabolic interactions. Within the plant microbiome, microbial communities use diverse mechanisms to thrive, survive, and multiply. In this review, Pacheco & Vorholt describe the interplay of metabolic interactions within plant microbiomes and review current…
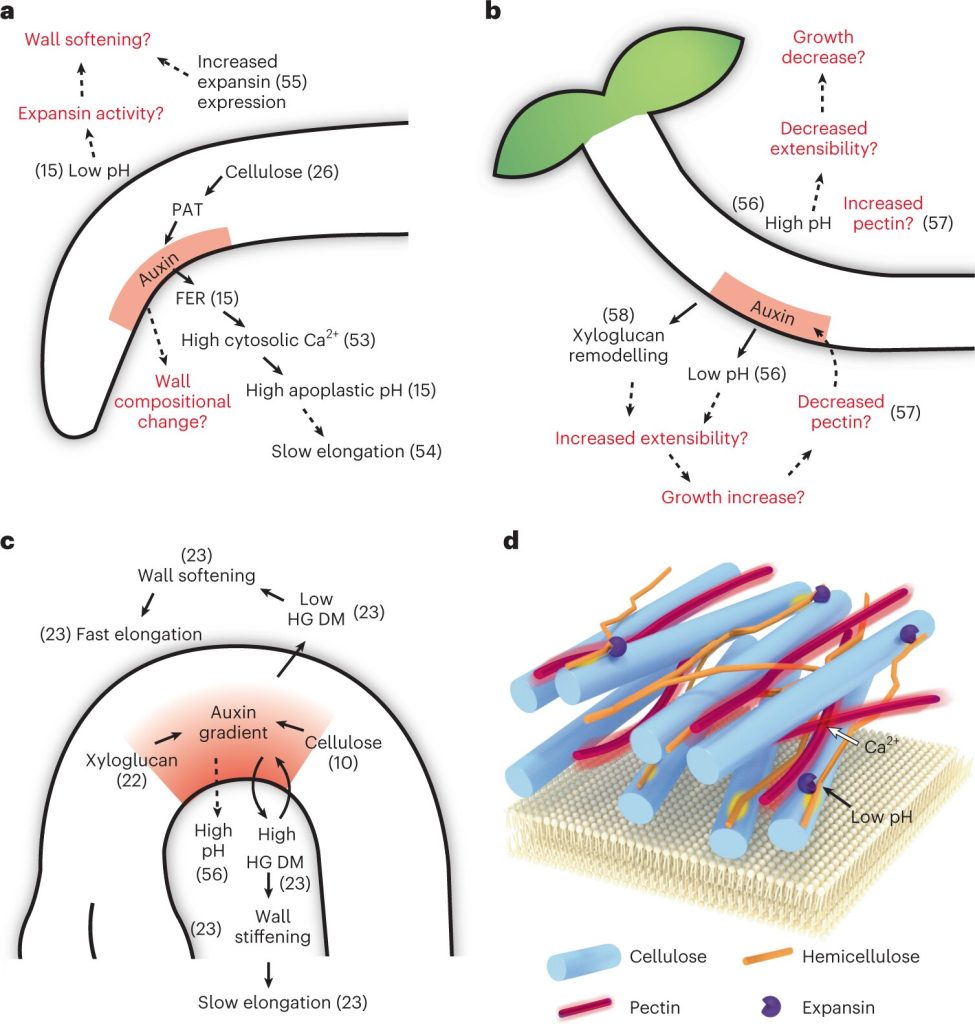
Perspective: Multiple mechanism behind plant bending
Plant Science Research WeeklyBending or folding in plants is influenced by different factors such as environmental conditions (nutrient, water, light, gravity), abiotic or biotic stress, cell wall properties, and cell differential growth. Jonsson et al. explain how bending is achieved by considering molecular mechanisms, mathematical…
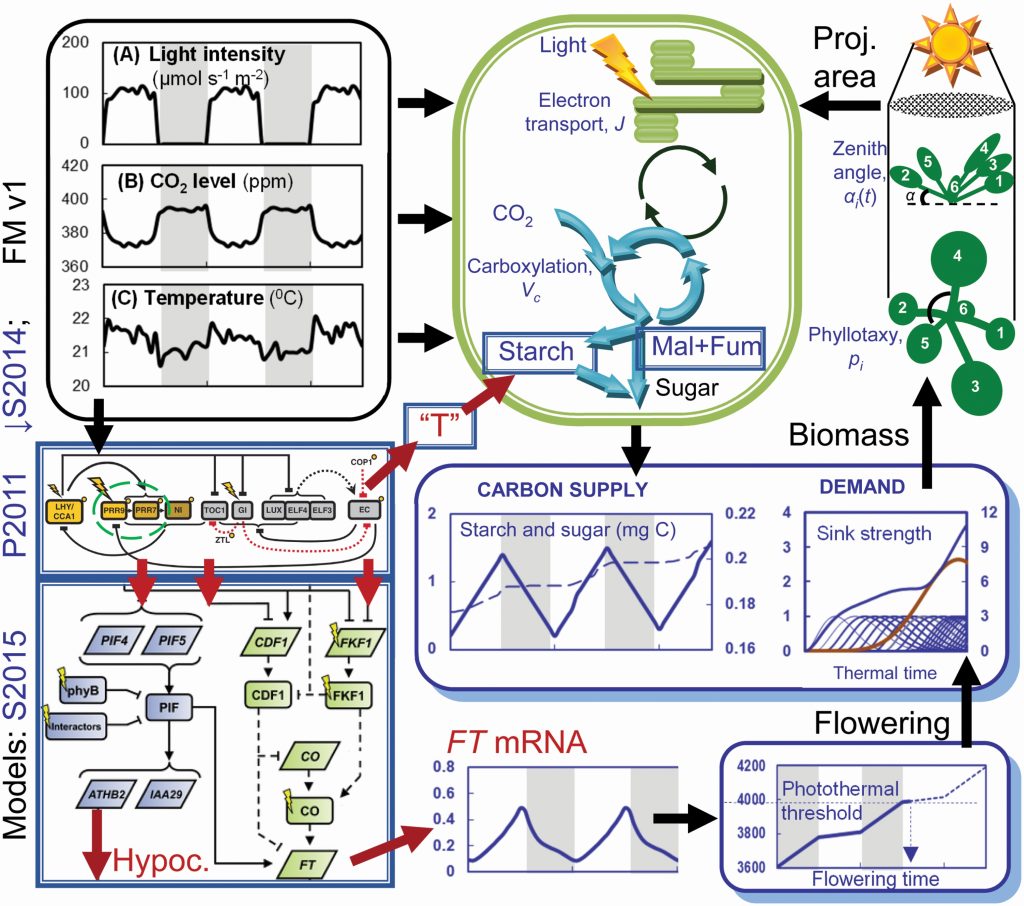
Arabidopsis Framework Model version 2 predicts the effects of circadian clock misregulation
Plant Science Research WeeklyExperimental or "wet lab" biologists look at data, develop a hypothesis to explain it, test the hypothesis, and repeat. Through these approaches, our understanding of life is continually being expanded and refined. A complementary approach involves developing mathematical models to explain observations,…

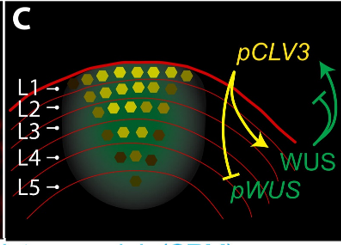
Concentration-dependent transcriptional switching through a collective action of cis-elements (Sci. Adv.)
Plant Science Research WeeklyOne of the well-known molecular pathways that regulate plant development is the one formed by the homeobox transcription factor WUSCHEL (WUS) and the secreted peptide CLAVATA 3 (CLV3). In the shoot apical meristem (SAM), WUS expression is constrained to the organizing center where it maintains the stem…

