
PeTriBERT : Augmenting BERT with tridimensional encoding for inverse protein folding and design (bioRxiv)
Plant Science Research WeeklyThe AlphaFoldDB database recently made headlines by predicting the three-dimensional protein structure of millions of proteins based on their primary amino acid sequence. Most of these models remain to be tested, but it’s a place to start. Here, Dumortier et al. add an important complementary tool…
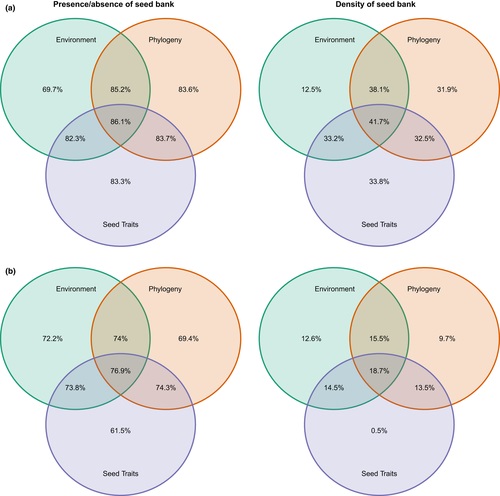
Machine learning algorithms predict soil seed bank persistence from easily available traits (Appl. Veg. Sci.)
Plant Science Research WeeklyKnowing whether a given plant species forms a persistent soil seed bank is essential to understanding its ecological dynamics, yet soil seed bank studies are both labor-intensive and time-consuming. One could argue that models should help predict this. Still, seed persistence in the soil is shaped by…
ATTED-II v11: A plant gene coexpression database using a sample balancing technique by subagging of principal components (Plant Cell Physiol)
Plant Science Research WeeklyGenes are generally expressed in concert with one another with data often represented by gene co-expression networks. Gene co-expression data may be useful in identifying genes with unknown functions and are generally utilized in exploring molecular processes occurring during normal development as well…
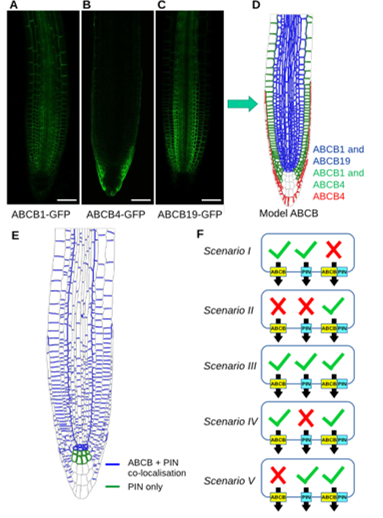
Systems approaches reveal that ABCB and PIN proteins mediate co-dependent auxin efflux (Plant Cell)
Plant Science Research WeeklyIn plant development, advanced computer model simulations allow the understanding of the non-intuitive relations between local morphogenetic processes and global patterning. In Arabidopsis thaliana, the hormone auxin is involved in several processes of plant development. Auxin transporters play an important…
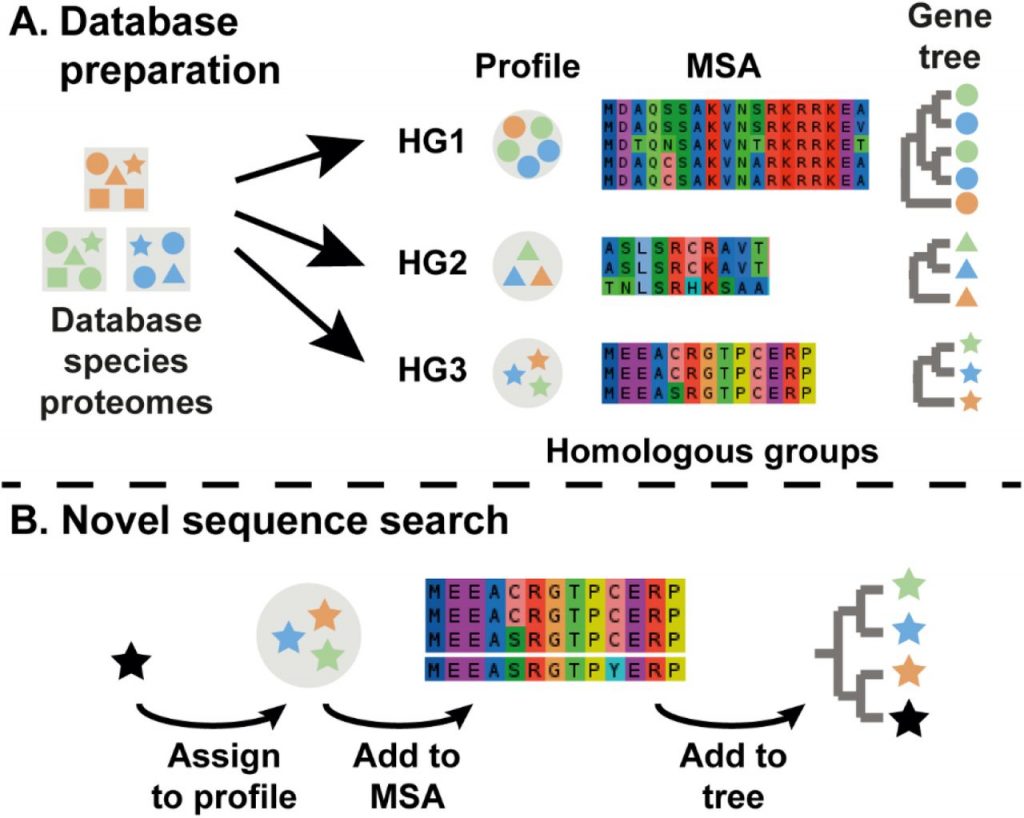
SHOOT: Phylogenetic gene search and ortholog inference (bioRxiv)
Plant Science Research WeeklyBLAST (basic local alignment search tool) is more than 30 years old, and most readers have probably had the opportunity to BLAST a sequence. BLAST takes an input sequence and searches databases for closely related sequences. However, this tool uses only sequence information to find close matches. Hypothetically,…
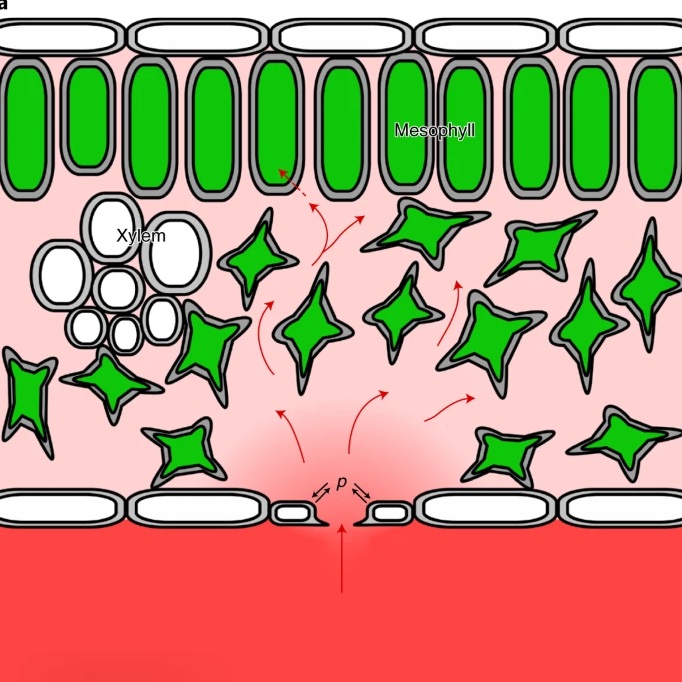
Guard cell endomembrane Ca2+-ATPases underpin a ‘carbon memory’ of photosynthetic assimilation that impacts on water-use efficiency (Nature Plants)
Plant Science Research WeeklyStomatal guard cells control both carbon dioxide uptake and transpirational water loss. Guard cell control over the stomatal pore aperture is sensitive to water status (through ABA) as well as the amount of CO2 available within the leaf air space. Several studies have indicated that stomatal aperture…
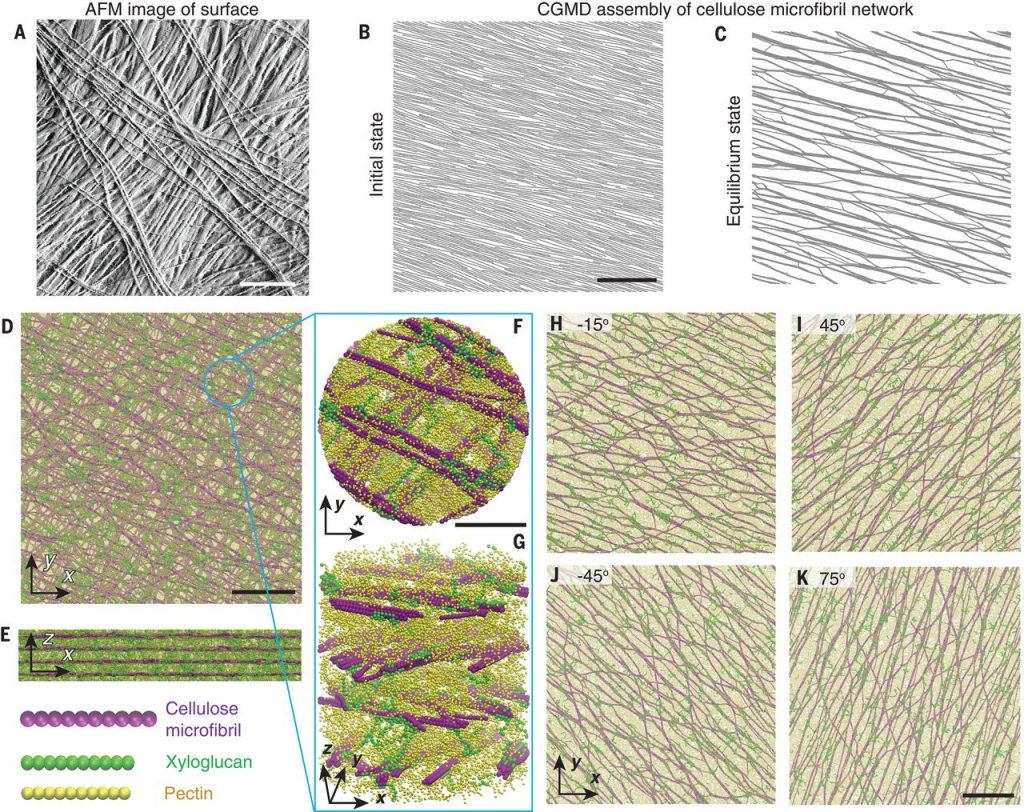
Molecular insights into the complex mechanics of plant epidermal cell walls (Science)
Plant Science Research WeeklyPrimary plant cell walls are made up of three types of polysaccharides: cellulose, hemicellulose, and pectin, the orientations and interactions of which provide the cell wall with its properties. Here, Zhang et al. examined the outer wall of onion epidermal cells through atomic-force microscopy, stress-strain…
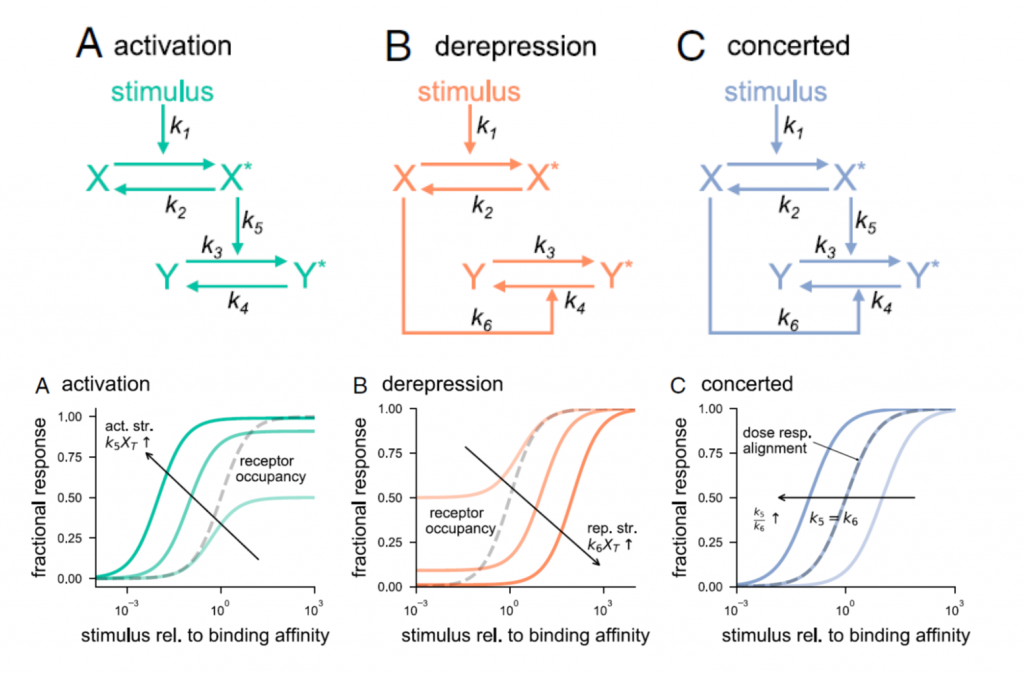
Molecular switch architecture determines response properties of signaling pathways (PNAS)
Plant Science Research WeeklyGenetic studies have provided us with countless examples of regulatory switches that transduce a signal into a response. Mutant analysis is usually sufficient to identify these controlling elements, but often in an all-or-nothing way. Here, Ghusinga et al. have taken a theoretical kinetic approach to…
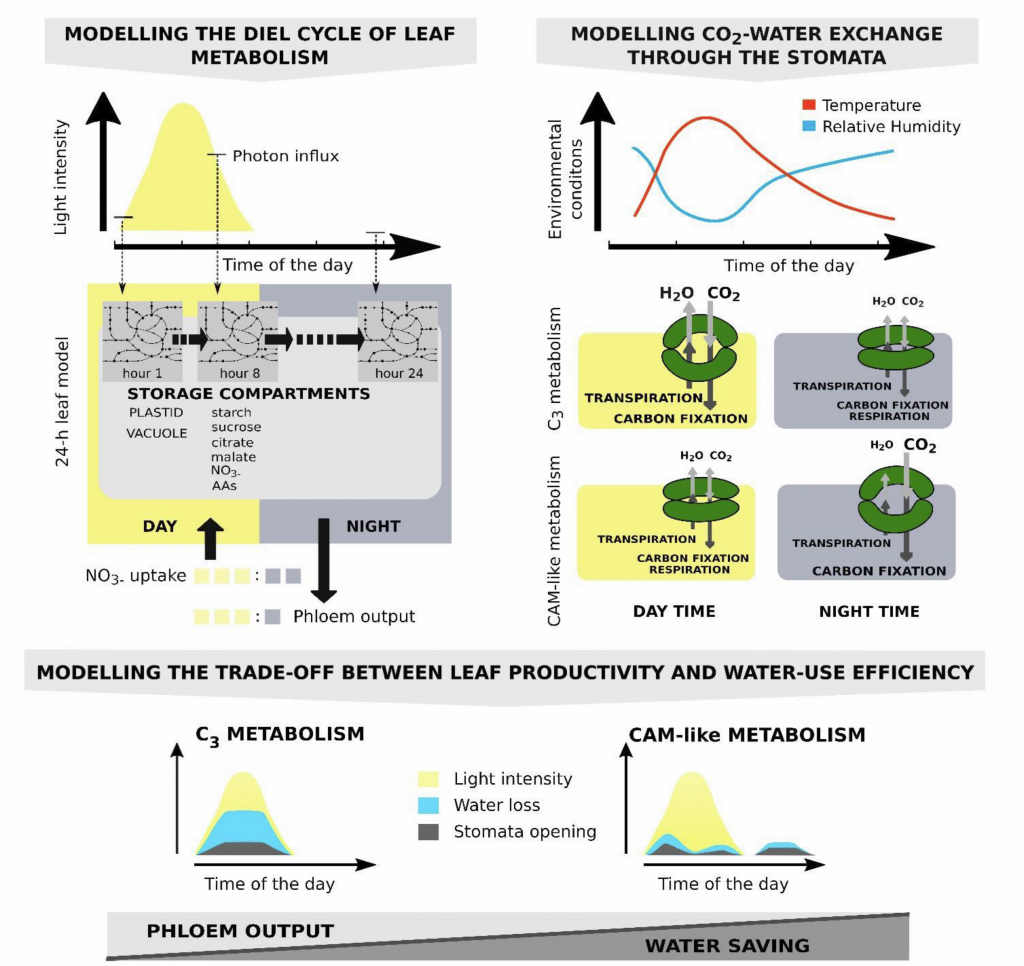
Alternative CAM and water-saving flux modes into C3 leaf metabolic model (Plant Cell)
Plant Science Research WeeklyCrassulacean acid metabolism (CAM) is a photosynthetic adaptation pathway in arid environments to minimize water loss by opening the stomata at night, when the temperature and therefore water loss due to transpiration is lower. Carbon dioxide is initially fixed at night and stored in the vacuole. Engineering…

