Identifying candidates from genome wide association studies using gene orthologs
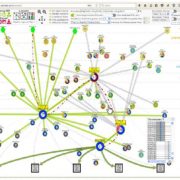
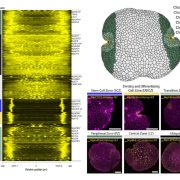
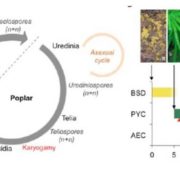
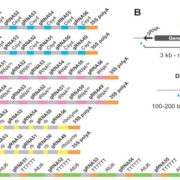
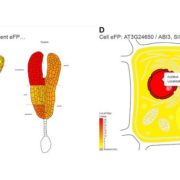
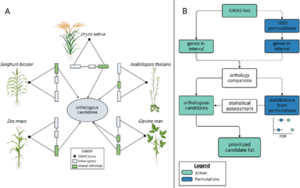 Genome wide association studies (GWAS) identify genomic loci associated with a specific trait. However, these loci often contain many genes, so selecting which to investigate further can be tricky. To improve this Whitt, et al. developed a program called FiReMAGE (filtering results of multi-species, analogous GWAS experiments), which takes loci from many GWAS experiments conducted in different species, identifies genes under each locus, and finds orthologs. Candidate genes are those where the ortholog group is detected in multiple species. Candidate prioritization follows running the pipeline with 1000 random loci and calculating the probability that the locus overlaps with the ortholog by chance. To demonstrate the potential of this program, the authors inputted metal accumulation GWAS data from Arabidopsis thaliana, soybean (Glycine max), rice (Oryza sativa), Sorghum bicolor and maize (Zea mays). By imposing the restriction that the ortholog group had to be identified in all five species, they found 81 strong candidates. Some of these have known roles in metal accumulation, such as the molybdate transport MOT2, whilst others are novel candidates, such as a mevalonate kinase associated with iron homeostasis. Hence, this program provides a powerful way of identifying gene candidates from GWAS loci. (Summary by Rose McNelly @Rose_McN) bioRxiv https://doi.org/10.1101/2023.10.05.561051
Genome wide association studies (GWAS) identify genomic loci associated with a specific trait. However, these loci often contain many genes, so selecting which to investigate further can be tricky. To improve this Whitt, et al. developed a program called FiReMAGE (filtering results of multi-species, analogous GWAS experiments), which takes loci from many GWAS experiments conducted in different species, identifies genes under each locus, and finds orthologs. Candidate genes are those where the ortholog group is detected in multiple species. Candidate prioritization follows running the pipeline with 1000 random loci and calculating the probability that the locus overlaps with the ortholog by chance. To demonstrate the potential of this program, the authors inputted metal accumulation GWAS data from Arabidopsis thaliana, soybean (Glycine max), rice (Oryza sativa), Sorghum bicolor and maize (Zea mays). By imposing the restriction that the ortholog group had to be identified in all five species, they found 81 strong candidates. Some of these have known roles in metal accumulation, such as the molybdate transport MOT2, whilst others are novel candidates, such as a mevalonate kinase associated with iron homeostasis. Hence, this program provides a powerful way of identifying gene candidates from GWAS loci. (Summary by Rose McNelly @Rose_McN) bioRxiv https://doi.org/10.1101/2023.10.05.561051