The Marchantia transcription factor atlas
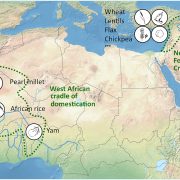
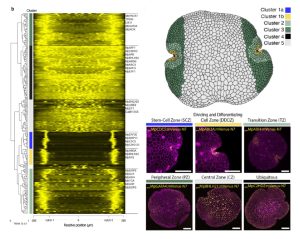 Marchantia’s power as a model organism continues to grow! Here, Ramoni et al. have investigated the expression pattern of the proximal promoters of most of its 450 transcription factor (TF)-encoding genes. The promoter elements were fused to nuclear-localized fluorescent reporters and introduced into plants. The authors provide the expression data of the constructs in a useful and accessible website, https://mpexpatdb.org/. Each TF promoter construct is shown along with chlorophyll fluorescence and a constitutive plasma membrane marker, which can be toggled on and off independently. Focusing on 218 genes, they identified five expression-pattern based clusters, with many of the genes preferentially expressed in the stem-cell zone or the dividing and differentiating cell zone. They also examined cell-specific and dynamic patterns of expression, and correlated these results with those obtained through other methods such as transcriptomics. As well as providing a great resource for further studies, this work has already provided insights into Marchantia development, including a suite of genes expressed in the stem-cell zone that are distinct from genes expressed in vascular plant meristems. (Summary by Mary Williams @PlantTeaching) Plant Cell 10.1093/plcell/koae053
Marchantia’s power as a model organism continues to grow! Here, Ramoni et al. have investigated the expression pattern of the proximal promoters of most of its 450 transcription factor (TF)-encoding genes. The promoter elements were fused to nuclear-localized fluorescent reporters and introduced into plants. The authors provide the expression data of the constructs in a useful and accessible website, https://mpexpatdb.org/. Each TF promoter construct is shown along with chlorophyll fluorescence and a constitutive plasma membrane marker, which can be toggled on and off independently. Focusing on 218 genes, they identified five expression-pattern based clusters, with many of the genes preferentially expressed in the stem-cell zone or the dividing and differentiating cell zone. They also examined cell-specific and dynamic patterns of expression, and correlated these results with those obtained through other methods such as transcriptomics. As well as providing a great resource for further studies, this work has already provided insights into Marchantia development, including a suite of genes expressed in the stem-cell zone that are distinct from genes expressed in vascular plant meristems. (Summary by Mary Williams @PlantTeaching) Plant Cell 10.1093/plcell/koae053