Back to where it came from: chloroplast expression of both Rubisco subunits helps functional enzyme analysis
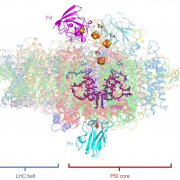
Rubisco catalyzes the key carboxylation step in photosynthetic CO2 fixation and is probably the most abundant protein on Earth. The enzyme is famous for inefficient catalysis and the habit of binding oxygen instead of CO2 in one out of every four binding events, leading to photorespiration reactions that release fixed CO2 and consume energy (Sharkey, 1988). Engineering an improved Rubisco to boost photosynthesis and increase crop yields is challenging, as interactions with Rubisco activase (required for enzyme activation) and chaperones (contributing to Rubisco assembly) need to be maintained (Sharwood, 2017). Further, coordinated changes in both the large (L) and small (S) subunits that make up the enzyme are needed to enhance its catalytic activity. Studying the role of various mutations on Rubisco activity in planta is hindered by the different genomic locations of genes encoding enzyme subunits: the rbcL gene encoding the Rubisco large subunit resides in the chloroplast genome, whereas multiple nuclear RBCS genes code for the small subunits. Now, Martin-Avila et al. (2020) have tackled this issue in tobacco (Nicotiana tabacum) by silencing the nuclear RBCS genes and introducing various rbcL and rbcS versions into the chloroplast genome to assess their function.
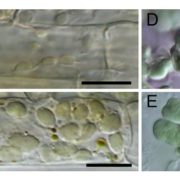
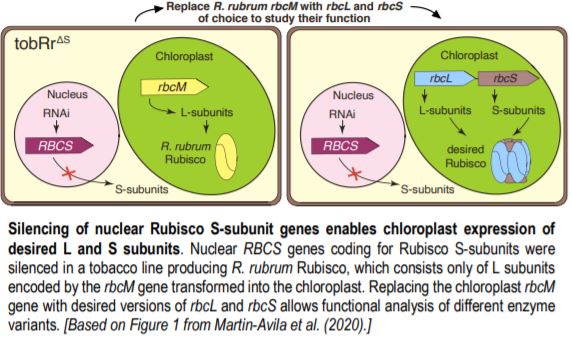 To avoid detrimental effects on photosynthesis and plant growth from impaired Rubisco function, the authors silenced the nuclear RBCS genes in a tobacco line carrying the gene for the Rubisco large subunit from Rhodospirillum rubrum (rbcM) in place of tobacco rbcL. The R. rubrum enzyme does not require S subunits; the obtained plants with 13 silenced RBCS genes, named tobRrΔS, were indistinguishable from the parental line.
To avoid detrimental effects on photosynthesis and plant growth from impaired Rubisco function, the authors silenced the nuclear RBCS genes in a tobacco line carrying the gene for the Rubisco large subunit from Rhodospirillum rubrum (rbcM) in place of tobacco rbcL. The R. rubrum enzyme does not require S subunits; the obtained plants with 13 silenced RBCS genes, named tobRrΔS, were indistinguishable from the parental line.
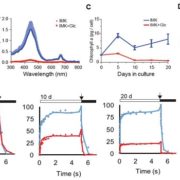
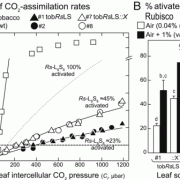
After successfully silencing endogenous RBCS genes, the authors used the tobRrΔS tobacco line to test the effect of different sequence features on Rubisco production from synthetic rbcL-rbcS operons transformed into the chloroplast genome to replace R. rubrum rbcM. They discovered that codon optimization was important to achieve high translation in the chloroplast, and thus high Rubisco levels. In addition, sequences around the translation initiation site for rbcS, the second gene in the operon, also strongly affected Rubisco production. The maximal Rubisco amounts obtained in this experiment from chloroplast expression only reached ~50% of wild type levels, which agrees with the slower plant growth and photosynthesis observed in the transplastomic plants.
The tobRrΔS is a great new tool to analyze Rubisco traits via expression of heterologous or mutant L and S subunits from the chloroplast genome (see Figure). The authors transformed the genes encoding the L subunit and one of four different S subunits from potato (Solanum tuberosum) Rubisco into the tobRrΔS chloroplast genome and studied the effect of each L-S combination on Rubisco biogenesis, kinetics, photosynthesis and plant growth. All combinations of chloroplast-expressed potato Rubisco subunits in tobacco yielded less enzyme and poorer photosynthesis and growth than in wild-type tobacco or in a line expressing potato L-subunit and native tobacco S-subunits. Nevertheless, the experiments gave insight into the structural characteristics that affect Rubisco activity. For example, the potato S3 subunit gave Rubisco a 9% higher catalytic activity compared to other potato S subunits, which was associated with two amino acid substitutions in the βA-βB loop in S3 that interacts with the L-subunit and may affect the interaction of the substrate with the catalytic site further down in the L-subunit.
The work by Martin-Avila et al. (2020) creates a valuable tool to study the effects of different substitutions in Rubisco subunits on enzyme biogenesis and catalytic activity, contributing to efforts of engineering a better Rubisco. However, optimization of mRNA biosynthesis and translation from transgenes incorporated into the chloroplast genome, as well as co-engineering Rubisco with the chaperones contributing to its assembly, are likely required to achieve higher Rubisco levels in transplastomic plants.
Hanna Hõrak
Institute of Technology
University of Tartu, Estonia
ORCID: 0000-0002-6392-859X
REFERENCES
Martin-Avila, E., Lim, Y., Birch, R., Dirk, L.M.A., Buck, S., Rhodes, T., Sharwood, R., Kapralov, M.V. and Whitney, S.M. (2020). Modifying plant photosynthesis and growth via simultaneous chloroplast transformation of Rubisco large and small subunits. Published July 9 2020. DOI: https://doi.org/10.1105/tpc.20.00288
Sharkey, T.D. (1988). Estimating the rate of photorespiration in leaves. Physiol. Plant. 73: 147–152.
Sharwood, R.E. (2017). Engineering chloroplasts to improve Rubisco catalysis: prospects for translating improvements into food and fiber crops. New Phytol. 213: 494–510.