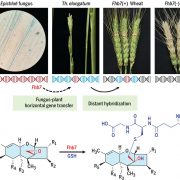
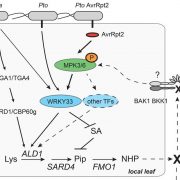
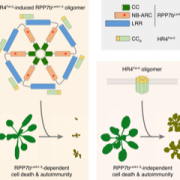
Active DNA demethylation controls defense gene regulation and antibacterial resistance (bioRxiv)
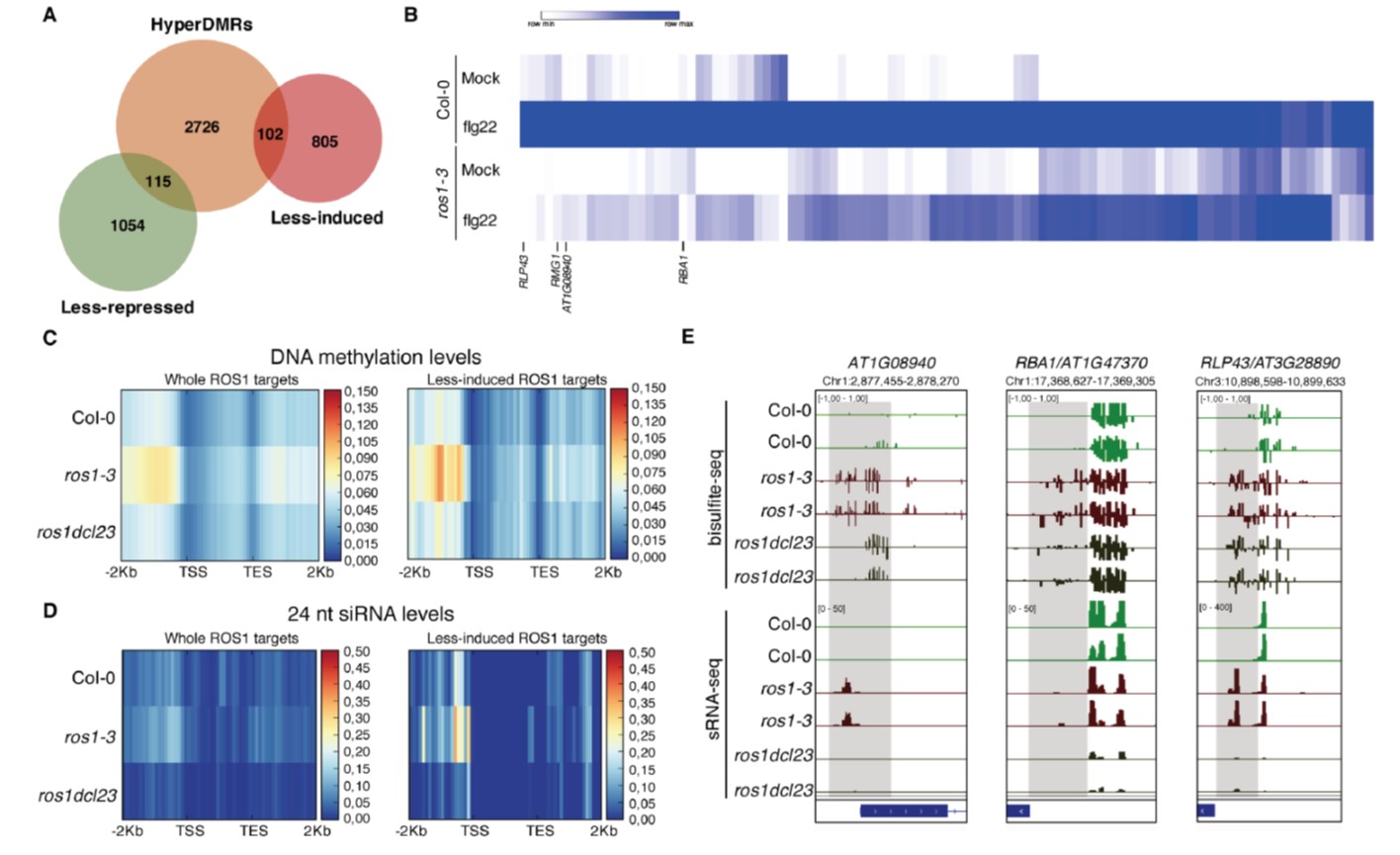
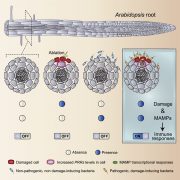
Epigenetic reprogramming, such as DNA methylation changes, has emerged as a crucial regulator of plant defense responses. Homeostasis of DNA methylation is controlled by a balance between methylation and demethylation. ROS1 (Repressor Of Silencing 1) is an Arabidopsis demethylases that is known to be a positive regulator of defense, but little is known about the mechanisms involved in this process. Halter et al. showed that ROS1 positively regulates plant defense against bacterial pathogens by antagonizing the function of DCL2/3 (DICER-LIKE 2/3), which play key roles in RNA-mediated DNA methylation (RdDM). A genome-wide analysis identified >2000 genes differentially regulated by ROS1 upon treatment of the immune elicitor flg22, among which 10% are demethylated by ROS1. Among such demethylated regions were WRKY transcription factor (TF) binding sites. The authors showed that hypermethylation at the promoter region of RLP43 (Receptor-Like Protein 43), a flg22 inducible gene, blocks binding of WRKY TFs to this DNA region. Furthermore, they showed that DNA demethylation in the promoter regions of defense-related genes, including RLP43, is important for the induction of these genes upon flg22 treatment and resistance against the bacterial pathogen Pseudomonas syringae. Taken together, this study reveals the molecular mechanisms underlying how ROS1-mediated demethylation plays critical roles in regulating plant defense responses and antibacterial resistance. (Summary by Tatsuya Nobori @nobolly) bioRxiv 10.1101/2020.09.30.321257