Extensive transcriptomic and epigenomic remodelling occurs during Arabidopsis thaliana germination
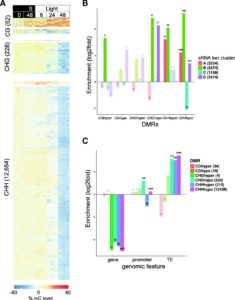 Understanding the complex regulatory mechanisms that contribute to germination and seedling establishment requires integration of gene-expression, transcription factors (TFs), DNA methylation, smallRNA data, and their interactions. Narsai et al. describe the first dynamic transcription factor network model of Arabidopsis seed germination over an extensive time-course, showing widespread alternative splicing and changes in known and novel regulatory genes. Their results uncovered both known germination regulators and novel TFs that control specific stages of germination using Dynamic Regulatory Events Miner (DREM) modeling of time course transcriptome data. The authors validate their results by showing delayed germination in T-DNA insertion lines carrying mutations in transcription factor genes identified using DREM. For the majority of these TFs, their ability to bind DNA is negatively affected by DNA methylation, shaping the regulatory network during vegetative growth. The authors also show significant changes in both miRNA and siRNA expression during seed germination and the transition to seedlings. They speculate whether miRNAs mediate DNA methylation changes by targeting genes involved in DNA methylation like DRM2 and IDNL-2. The epigenome undertoes extensive demethylation during the transition from embryo to seedling and there is significant overlap between CHH-hypomethylated DMRs and siRNA clusters, indicating that the decrease in CHH methylation is likely due to the RNA-directed DNA Methylation pathway (RdDM) and not the CMT2-mediated pathway. Genome Biology 10.1186/s13059-017-1302-3
Understanding the complex regulatory mechanisms that contribute to germination and seedling establishment requires integration of gene-expression, transcription factors (TFs), DNA methylation, smallRNA data, and their interactions. Narsai et al. describe the first dynamic transcription factor network model of Arabidopsis seed germination over an extensive time-course, showing widespread alternative splicing and changes in known and novel regulatory genes. Their results uncovered both known germination regulators and novel TFs that control specific stages of germination using Dynamic Regulatory Events Miner (DREM) modeling of time course transcriptome data. The authors validate their results by showing delayed germination in T-DNA insertion lines carrying mutations in transcription factor genes identified using DREM. For the majority of these TFs, their ability to bind DNA is negatively affected by DNA methylation, shaping the regulatory network during vegetative growth. The authors also show significant changes in both miRNA and siRNA expression during seed germination and the transition to seedlings. They speculate whether miRNAs mediate DNA methylation changes by targeting genes involved in DNA methylation like DRM2 and IDNL-2. The epigenome undertoes extensive demethylation during the transition from embryo to seedling and there is significant overlap between CHH-hypomethylated DMRs and siRNA clusters, indicating that the decrease in CHH methylation is likely due to the RNA-directed DNA Methylation pathway (RdDM) and not the CMT2-mediated pathway. Genome Biology 10.1186/s13059-017-1302-3