DNA methylation dynamics during early plant life
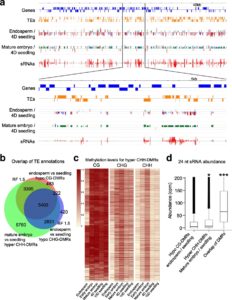 DNA methylation is extensively reprogrammed in the early embryo and germlines of mammals, whereas flowering plants do not show such extensive resetting except in the endosperm. Active DNA demethylation in the central cell and reduced activity of DNA methyltransferases leads to global hypomethylation of the maternal genome in the endosperm. On the paternal side, the vegetative nucleus of the pollen undergoes active DNA demethylation, but this is limited to only a few transposable element (TE) sequences. Bouyer et al. used Arabidopsis to examine DNA methylation dynamics during embryogenesis and early vegetative development. They show higher non-CG methylation in the mature embryo compared to seedlings and adult plants. Methylation at CHH sites (mCHH) makes up 44% of all methylated cytosines in embryos, compared to less than 27% in seedlings, particularly in the pericentromeric regions. CHH hypermethylation in mature embryo coincides with the hypomethylated state during endosperm development and a higher abundance of 24nt siRNAs. Based on their analysis of temporal gene expression and patterns of mCHH on long TE sequences in the early and mature embryo, the authors suggest that siRNA production in the endosperm precedes RNA-directed DNA Methylation (RdDM) in the mature embryo. RdDM is active in every cell of the mature embryo in contrast to its activity being restricted to meristems post-embryonically. They also show that failure to terminate the embryonic developmental program as seen in fie mutants leads to massive CHH hypermethylation retained in the seedlings, particularly on RdDM targets. (Summary by Sunil Kumar) Genome Biology 10.1186/s13059-017-1313-0
DNA methylation is extensively reprogrammed in the early embryo and germlines of mammals, whereas flowering plants do not show such extensive resetting except in the endosperm. Active DNA demethylation in the central cell and reduced activity of DNA methyltransferases leads to global hypomethylation of the maternal genome in the endosperm. On the paternal side, the vegetative nucleus of the pollen undergoes active DNA demethylation, but this is limited to only a few transposable element (TE) sequences. Bouyer et al. used Arabidopsis to examine DNA methylation dynamics during embryogenesis and early vegetative development. They show higher non-CG methylation in the mature embryo compared to seedlings and adult plants. Methylation at CHH sites (mCHH) makes up 44% of all methylated cytosines in embryos, compared to less than 27% in seedlings, particularly in the pericentromeric regions. CHH hypermethylation in mature embryo coincides with the hypomethylated state during endosperm development and a higher abundance of 24nt siRNAs. Based on their analysis of temporal gene expression and patterns of mCHH on long TE sequences in the early and mature embryo, the authors suggest that siRNA production in the endosperm precedes RNA-directed DNA Methylation (RdDM) in the mature embryo. RdDM is active in every cell of the mature embryo in contrast to its activity being restricted to meristems post-embryonically. They also show that failure to terminate the embryonic developmental program as seen in fie mutants leads to massive CHH hypermethylation retained in the seedlings, particularly on RdDM targets. (Summary by Sunil Kumar) Genome Biology 10.1186/s13059-017-1313-0