Multi-omics analyses of wild tomato introgression lines reveal a set of gene expression-metabolite-pathogen sensitivity interactions (Nat. Genet.)
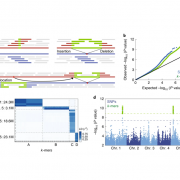
 Domesticated tomato has been subjected to human selection to satisfy marketing and economic desires, leading to a loss of genetic diversity and elimination of fruits traits such as flavor, aroma and pathogen resistance. Introgression lines (ILs) that capture the genome of wild desert-adapted tomato Solanum pennellii in the background of cultivated tomato, S. lycopersicum, provide a remarkable tool to improve the resolution of Quantitative Trait Locus (QTL) mapping. Szymański and colleagues carried out transcriptome-metabolome-pathogen sensitivity analysis in breaker (slightly pink) and red ripe stages of 580 backcrossed inbred lines (BILs) and ILs. The multi-omics analysis identified genomic loci associated with hundreds of transcripts and metabolites related to tomato fruits development and ripening. The analysis uncovered core enzyme components of the tomato alkaloid pathway as well as a set of genes and metabolites promoting fungal resistance, which highlight the role of phenylpropanoids in fruit defenses. This large dataset will be an resource finding for molecular breeding in selection for flavor and pathogen resistance and is accessible through an online data browser https://szymanskilab.shinyapps.io/kilbil/. (Summary by Min May Wong @wongminmay) Nat. Genet. 10.1038/s41588-020-0690-6
Domesticated tomato has been subjected to human selection to satisfy marketing and economic desires, leading to a loss of genetic diversity and elimination of fruits traits such as flavor, aroma and pathogen resistance. Introgression lines (ILs) that capture the genome of wild desert-adapted tomato Solanum pennellii in the background of cultivated tomato, S. lycopersicum, provide a remarkable tool to improve the resolution of Quantitative Trait Locus (QTL) mapping. Szymański and colleagues carried out transcriptome-metabolome-pathogen sensitivity analysis in breaker (slightly pink) and red ripe stages of 580 backcrossed inbred lines (BILs) and ILs. The multi-omics analysis identified genomic loci associated with hundreds of transcripts and metabolites related to tomato fruits development and ripening. The analysis uncovered core enzyme components of the tomato alkaloid pathway as well as a set of genes and metabolites promoting fungal resistance, which highlight the role of phenylpropanoids in fruit defenses. This large dataset will be an resource finding for molecular breeding in selection for flavor and pathogen resistance and is accessible through an online data browser https://szymanskilab.shinyapps.io/kilbil/. (Summary by Min May Wong @wongminmay) Nat. Genet. 10.1038/s41588-020-0690-6