Versatile auto-luminescent luciferase-based reporters for spatiotemporal gene expression studies (eLIFE)
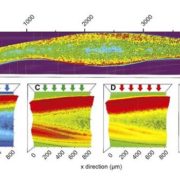
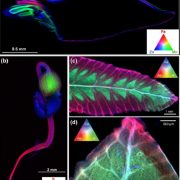
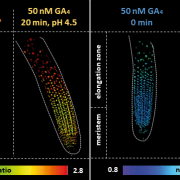
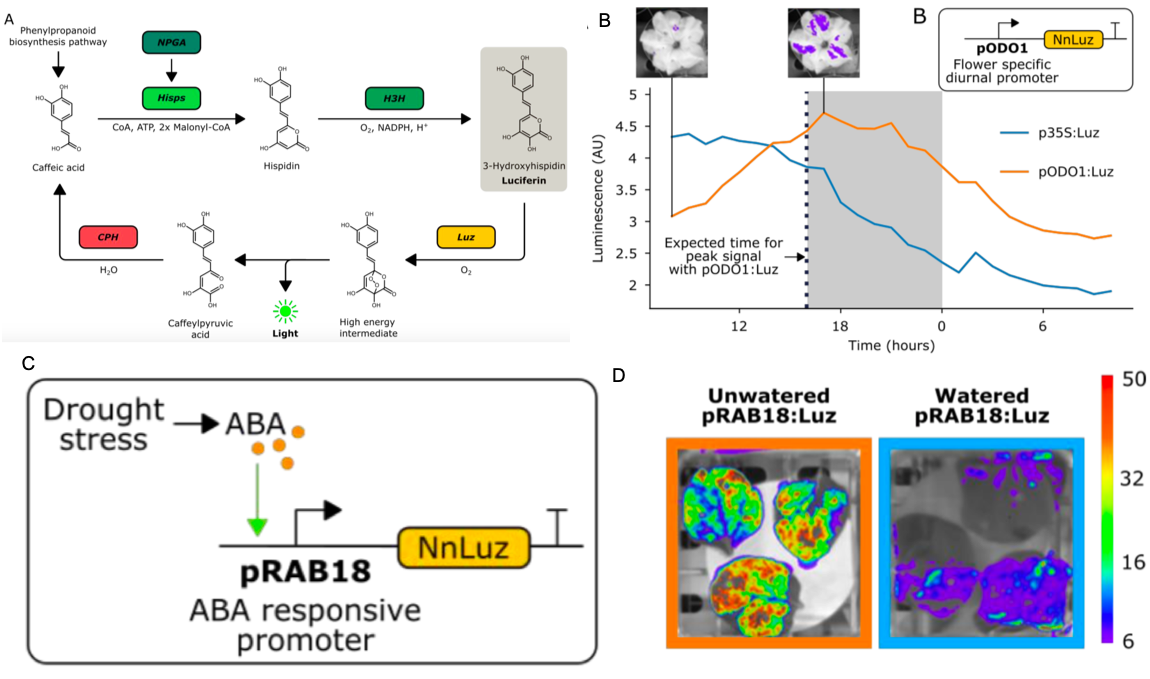 Firefly luciferase constructs are extensively used for gene expression studies but require a reaction with an exogenous substrate, so expression to some extent reflects tissue penetrance. Khakhar et al. designed customizable auto-luminescence constructs based on a fungal bioluminescence pathway (FBP) in which caffeic acid, an intermediate of lignin biosynthesis, is converted into luciferin by the action of three fungal enzymes. Auto-luminescence was observed following infiltration with Agrobacterium carrying the FBP constructs into Nicotiana leaves, with the luminescence enhanced by also introducing an enzyme that recycles the byproduct of the luciferase reaction or engineering increased expression of caffeic acid biosynthesis. These constructs can be modified to use in a variety of plant species like Arabidopsis, petunia, tomato, dahlia, periwinkle, and rose. As examples, using this system the authors observed a diurnal expression pattern from the ODORANT1 promoter in petunia and changes in transcription in response to abscisic acid flux in Arabidopsis. Other advantages are that the signal of the luciferase reaction is emitted in the green spectrum, making it compatible to use along with the conventional firefly luciferase assay, and imaging uses an entry-level DSLR camera along with Raspberry Pi single board computer instead of more expensive CCD cameras. These new tools offer an economic way to study gene expression changes over long periods during environmental and hormonal changes and can be used to study plant-pollinator interactions and many other creative ways. (Summary by Vijaya Batthula) eLife 10.7554/eLife.52786
Firefly luciferase constructs are extensively used for gene expression studies but require a reaction with an exogenous substrate, so expression to some extent reflects tissue penetrance. Khakhar et al. designed customizable auto-luminescence constructs based on a fungal bioluminescence pathway (FBP) in which caffeic acid, an intermediate of lignin biosynthesis, is converted into luciferin by the action of three fungal enzymes. Auto-luminescence was observed following infiltration with Agrobacterium carrying the FBP constructs into Nicotiana leaves, with the luminescence enhanced by also introducing an enzyme that recycles the byproduct of the luciferase reaction or engineering increased expression of caffeic acid biosynthesis. These constructs can be modified to use in a variety of plant species like Arabidopsis, petunia, tomato, dahlia, periwinkle, and rose. As examples, using this system the authors observed a diurnal expression pattern from the ODORANT1 promoter in petunia and changes in transcription in response to abscisic acid flux in Arabidopsis. Other advantages are that the signal of the luciferase reaction is emitted in the green spectrum, making it compatible to use along with the conventional firefly luciferase assay, and imaging uses an entry-level DSLR camera along with Raspberry Pi single board computer instead of more expensive CCD cameras. These new tools offer an economic way to study gene expression changes over long periods during environmental and hormonal changes and can be used to study plant-pollinator interactions and many other creative ways. (Summary by Vijaya Batthula) eLife 10.7554/eLife.52786
[altmetric doi=”10.7554/eLife.52786″ details=”right” float=”right”]