Review: Methods to visualize elements in plants (Plant Physiol)
Plant Science Research Weekly: February 14
Review: Deep learning for plant genomics and crop improvement
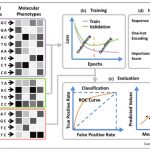
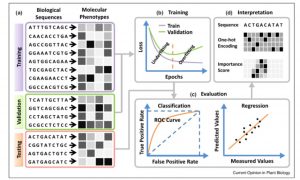 One of the goals of plant science is to use the molecular phenotype (genome, transcriptome, proteome) to predict the whole-plant phenotype. Deep learning approaches can potentially begin to do this, starting with a training dataset, and testing it with a validation dataset. Wang et al. review advances in towards this goal, particularly those that employ deep learning and neural networks, such as Convolutional Neural Networks (CNNs). The authors define different types of neural networks, and how they can be optimized for example to identify single nucleotide effects on traits, or on transcription factor binding. They also describe methods that have been developed to predict protein structure or protein-protein interactions. The authors discuss the possibilities and pitfalls of adapting deep learning models developed for animal or human genomes to plants, as well as plant-specific challenges such as polyplody. Plants’ phenotypic sensitivity to the environment adds additional complexity. The authors conclude that deep learning is a “promising approach for genome-wide identification of deleterious and adaptive variants, a prerequisite for editing-based genetic improvement of crops in future agriculture.” (Summary by Mary Williams) Curr. Opin. Plant Biol. 10.1016/j.pbi.2019.12.010
One of the goals of plant science is to use the molecular phenotype (genome, transcriptome, proteome) to predict the whole-plant phenotype. Deep learning approaches can potentially begin to do this, starting with a training dataset, and testing it with a validation dataset. Wang et al. review advances in towards this goal, particularly those that employ deep learning and neural networks, such as Convolutional Neural Networks (CNNs). The authors define different types of neural networks, and how they can be optimized for example to identify single nucleotide effects on traits, or on transcription factor binding. They also describe methods that have been developed to predict protein structure or protein-protein interactions. The authors discuss the possibilities and pitfalls of adapting deep learning models developed for animal or human genomes to plants, as well as plant-specific challenges such as polyplody. Plants’ phenotypic sensitivity to the environment adds additional complexity. The authors conclude that deep learning is a “promising approach for genome-wide identification of deleterious and adaptive variants, a prerequisite for editing-based genetic improvement of crops in future agriculture.” (Summary by Mary Williams) Curr. Opin. Plant Biol. 10.1016/j.pbi.2019.12.010
Review: Methods to visualize elements in plants
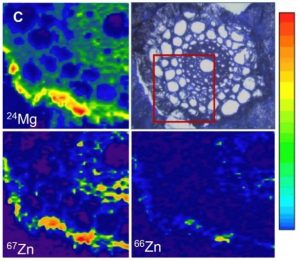
 How elements are taken up, transported, accumulated, and reach homeostasis within tissues are fundamental questions in plant molecular biology. Some aspects of plant mineral nutrition and crop productivity are related to element distribution: for instance, the analysis of fertilizer translocation upon application, or how plants can tolerate high concentrations of some metals in their tissues. The distribution and concentration of nutrients in fruits and seeds are vital for human health. In a recent review, Kopittke and colleagues summarize different techniques to study element distribution in plants. In the X-ray fluorescence-based approaches, a beam of high-energy X-rays, electrons, or protons is directed to the sample, exciting different elements and allowing their identification based upon their characteristic fluorescent X-rays. In the mass spectrometry-based approaches, small portions of the sample are progressively analyzed. One of the classic techniques is autoradiography; radioactive isotopes (e.g., 26Al, 33P, 35S, 45Ca, 54Mn, 55Fe, 67Cu) can be tracked throughout the plant tissues. Another technique for in vivo analysis is the use of laser confocal microscopy with element-selective fluorophores. The currently available fluorophores allow the detection of Zn, Ni/Co, Cu, or Pb/Cd. The authors provide a comprehensive comparison of those techniques, including key features such as: accessibility, maximum resolution, scan speed, detections limits, and the suitability for in vivo analysis. (Summary by Humberto Herrera-Ubaldo)
How elements are taken up, transported, accumulated, and reach homeostasis within tissues are fundamental questions in plant molecular biology. Some aspects of plant mineral nutrition and crop productivity are related to element distribution: for instance, the analysis of fertilizer translocation upon application, or how plants can tolerate high concentrations of some metals in their tissues. The distribution and concentration of nutrients in fruits and seeds are vital for human health. In a recent review, Kopittke and colleagues summarize different techniques to study element distribution in plants. In the X-ray fluorescence-based approaches, a beam of high-energy X-rays, electrons, or protons is directed to the sample, exciting different elements and allowing their identification based upon their characteristic fluorescent X-rays. In the mass spectrometry-based approaches, small portions of the sample are progressively analyzed. One of the classic techniques is autoradiography; radioactive isotopes (e.g., 26Al, 33P, 35S, 45Ca, 54Mn, 55Fe, 67Cu) can be tracked throughout the plant tissues. Another technique for in vivo analysis is the use of laser confocal microscopy with element-selective fluorophores. The currently available fluorophores allow the detection of Zn, Ni/Co, Cu, or Pb/Cd. The authors provide a comprehensive comparison of those techniques, including key features such as: accessibility, maximum resolution, scan speed, detections limits, and the suitability for in vivo analysis. (Summary by Humberto Herrera-Ubaldo)