Genome-phenome wide association in maize and Arabidopsis identifies a common molecular and evolutionary signature (Mol. Plant)
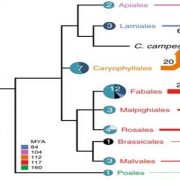
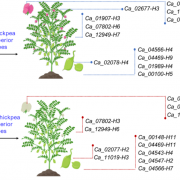
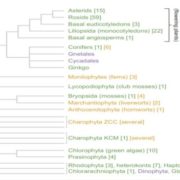
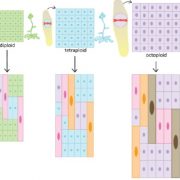
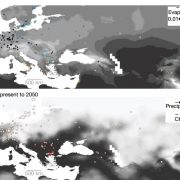
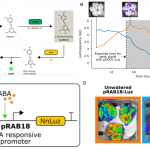
 Genome-wide association studies (GWAS) are widely used to link natural genetic variation to trait variation, with a single or a select few correlated traits assessed. High-throughput phenotyping allows the scoring of hundreds of individuals for various traits at several time points. An undeniable consequence of quantitative genetics research is the creation of large, complex datasets that can contain information on a multitude of traits in a population. Here, Liang et al. conducted a Genome-Phenome Wide Association Study (GPWAS) on maize and Arabidopsis. While a conventional GWAS only considers traits that are correlated, this novel GPWAS approach links gene markers to several correlated and uncorrelated traits simultaneously. The authors found that genes identified using this “multi-trait and multi-SNP framework” are more similar to those previously characterized using forward genetics approaches than genes from annotated gene models or conventional GWAS. GPWAS-identified maize genes resemble those with verified loss-of-function phenotypes at the molecular, population, and evolutionary levels by higher expression levels, syntenic conservation, and stronger purifying selection respectively, with similar observations made for Arabidopsis. Interestingly, genes linked to variation in individual phenotypes by GWAS had intermediate values between the annotated gene models and GPWAS/classical mutants. This may suggest that GPWAS is more likely to detect pleiotropic genes while conventional GWAS is more suitable for identifying loci that control a smaller number of phenotypes. Nonetheless, GPWAS represents an interesting approach for using the wealth of data available for model organisms such as maize, rice, and Arabidopsis. (Summary by Caroline Dowling) Mol. Plant 10.1016/j.molp.2020.03.003
Genome-wide association studies (GWAS) are widely used to link natural genetic variation to trait variation, with a single or a select few correlated traits assessed. High-throughput phenotyping allows the scoring of hundreds of individuals for various traits at several time points. An undeniable consequence of quantitative genetics research is the creation of large, complex datasets that can contain information on a multitude of traits in a population. Here, Liang et al. conducted a Genome-Phenome Wide Association Study (GPWAS) on maize and Arabidopsis. While a conventional GWAS only considers traits that are correlated, this novel GPWAS approach links gene markers to several correlated and uncorrelated traits simultaneously. The authors found that genes identified using this “multi-trait and multi-SNP framework” are more similar to those previously characterized using forward genetics approaches than genes from annotated gene models or conventional GWAS. GPWAS-identified maize genes resemble those with verified loss-of-function phenotypes at the molecular, population, and evolutionary levels by higher expression levels, syntenic conservation, and stronger purifying selection respectively, with similar observations made for Arabidopsis. Interestingly, genes linked to variation in individual phenotypes by GWAS had intermediate values between the annotated gene models and GPWAS/classical mutants. This may suggest that GPWAS is more likely to detect pleiotropic genes while conventional GWAS is more suitable for identifying loci that control a smaller number of phenotypes. Nonetheless, GPWAS represents an interesting approach for using the wealth of data available for model organisms such as maize, rice, and Arabidopsis. (Summary by Caroline Dowling) Mol. Plant 10.1016/j.molp.2020.03.003
[altmetric doi=”10.1016/j.molp.2020.03.003″ details=”right” float=”right”]