Extensive genome study to boost yield and improve agronomic traits in chickpea (Nature)
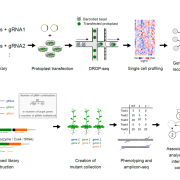
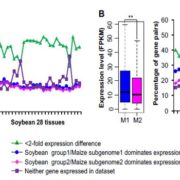
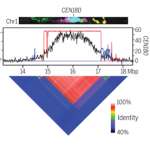
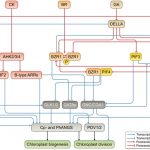
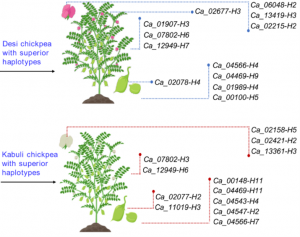 In recent years, there has been increasing awareness about the environmental impact of animal-based protein sources. Legumes such as chickpea (Cicer arientinum) are relatively cheap and sustainable sources of proteins, dietary fibres and micronutrients. Although there is a vast chickpea germplasm collection, little genomic characterization has been done. In Varshney et al., an international team of researchers from 41 organizations assembled the chickpea pan-genome by sequencing 3,366 accessions, includeing 3,171 cultivated and 195 wild varieties. A total of 29,870 genes, including 1582 novel genes, were identified in this study. Based on gene ontology analysis of the novel genes, several genes involved in adaptation to abiotic stresses have been identified. Further, the authors predicted the divergence and migration pattern of Cicer species across the globe based on the homology-based gene annotation. Most importantly, this study identified deleterious mutations and candidate genes responsible for reduced genetic diversity and fitness. The authors also report superior haplotypes (sets of DNA variations that tend not to recombine, and therefore are passed down through the generations together) in the local chickpea varieties that have distinctive characteristics arising from development and adaptation in a particular geographical area. This study provides a rich genetic resource for chickpea improvement and suggests ways to produce climate resilient and nutrient dense crops. (Summary by Chandan Kumar Gautam @chandan_gautam) Nature 10.1038/s41586-021-04066-1
In recent years, there has been increasing awareness about the environmental impact of animal-based protein sources. Legumes such as chickpea (Cicer arientinum) are relatively cheap and sustainable sources of proteins, dietary fibres and micronutrients. Although there is a vast chickpea germplasm collection, little genomic characterization has been done. In Varshney et al., an international team of researchers from 41 organizations assembled the chickpea pan-genome by sequencing 3,366 accessions, includeing 3,171 cultivated and 195 wild varieties. A total of 29,870 genes, including 1582 novel genes, were identified in this study. Based on gene ontology analysis of the novel genes, several genes involved in adaptation to abiotic stresses have been identified. Further, the authors predicted the divergence and migration pattern of Cicer species across the globe based on the homology-based gene annotation. Most importantly, this study identified deleterious mutations and candidate genes responsible for reduced genetic diversity and fitness. The authors also report superior haplotypes (sets of DNA variations that tend not to recombine, and therefore are passed down through the generations together) in the local chickpea varieties that have distinctive characteristics arising from development and adaptation in a particular geographical area. This study provides a rich genetic resource for chickpea improvement and suggests ways to produce climate resilient and nutrient dense crops. (Summary by Chandan Kumar Gautam @chandan_gautam) Nature 10.1038/s41586-021-04066-1