Unraveling cis and trans regulatory evolution during cotton domestication (Nature Comms)
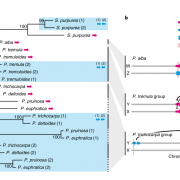
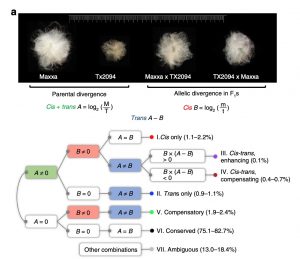 Polyploidization leads to a myriad of changes in gene expression and organization of genomes and can supply the material for speciation, adaptation, and morphological innovation. The most cultivated cotton species, Gossypium hirsutum, is an allotetraploid species (AD genome) containing two subgenomes (A from African and D from American diploid progenitors). Bao et al. have analyzed 27 RNA-seq libraries of four G. hirsutum accessions (wild and domesticated parental cotton and their reciprocal F1 hybrids) in two fiber developmental stages (fibers are single-celled seed trichomes). Through this they identified potential cis- and trans-regulatory element variation during cotton domestication, their inheritance patterns, and their association in co-expression and regulatory networks that are imperative for fiber development. The findings described by the study enlighten the evolutionary story of cotton fiber, and they will be very useful for agricultural improvement strategies in G. hirsutum and other polyploid species. (Summary by Sarah M. Nardeli) Nature Comms. 10.1038/s41467-019-13386-w
Polyploidization leads to a myriad of changes in gene expression and organization of genomes and can supply the material for speciation, adaptation, and morphological innovation. The most cultivated cotton species, Gossypium hirsutum, is an allotetraploid species (AD genome) containing two subgenomes (A from African and D from American diploid progenitors). Bao et al. have analyzed 27 RNA-seq libraries of four G. hirsutum accessions (wild and domesticated parental cotton and their reciprocal F1 hybrids) in two fiber developmental stages (fibers are single-celled seed trichomes). Through this they identified potential cis- and trans-regulatory element variation during cotton domestication, their inheritance patterns, and their association in co-expression and regulatory networks that are imperative for fiber development. The findings described by the study enlighten the evolutionary story of cotton fiber, and they will be very useful for agricultural improvement strategies in G. hirsutum and other polyploid species. (Summary by Sarah M. Nardeli) Nature Comms. 10.1038/s41467-019-13386-w