Review: Harnessing the power of gene regulatory networks for crop improvement
Plant Science Research WeeklyThe number of large molecular plant datasets seems to be growing exponentially. With this growth comes an urgent need for researchers to understand how these datasets should be analyzed and integrated to get accurate understandings of what they represent. This fine article by Leong et al. is a good place…
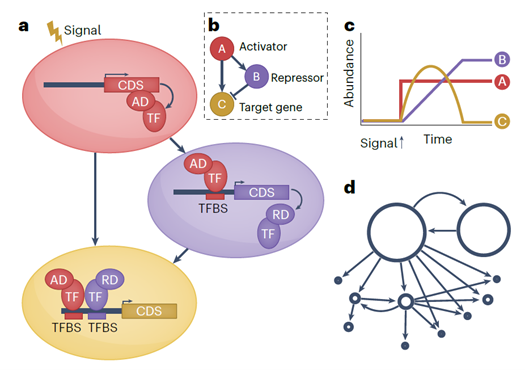
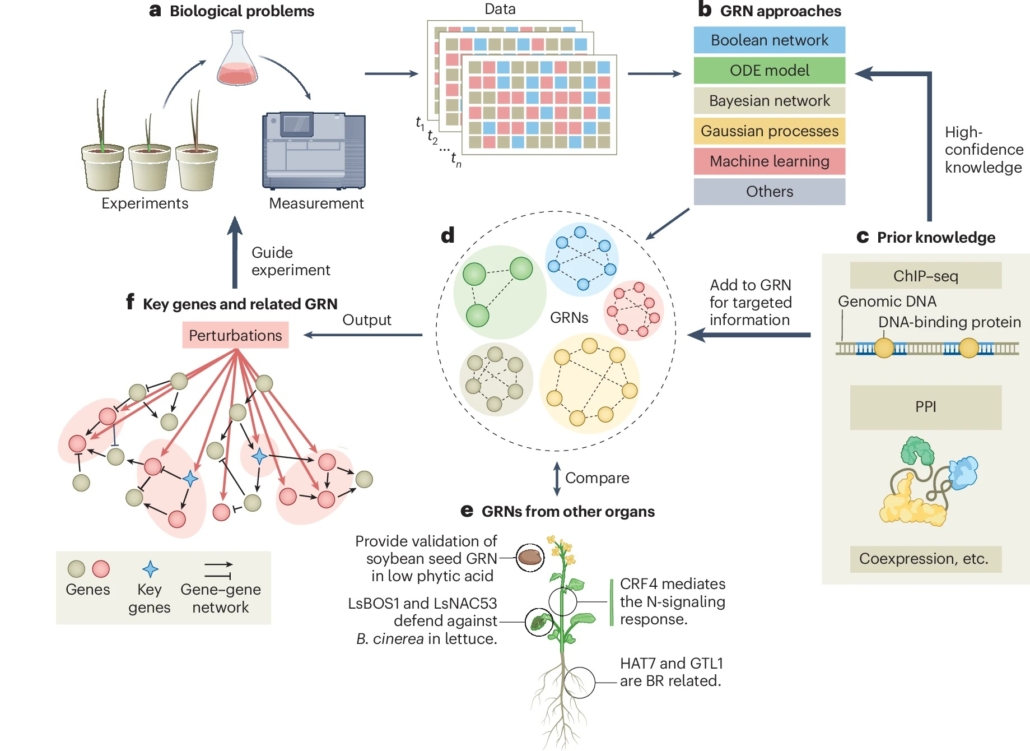
Review: Defining and rewiring of gene regulatory networks for plant improvement
Plant Science Research WeeklyMuch of the focus of functional genomics studies in plants is to improve yield, disease resistance, abiotic stress tolerance and nutritional quality. Many desirable traits are controlled by sets of genes that are coordinated in a complex network, called a gene regulatory network (GRN). A transcription…
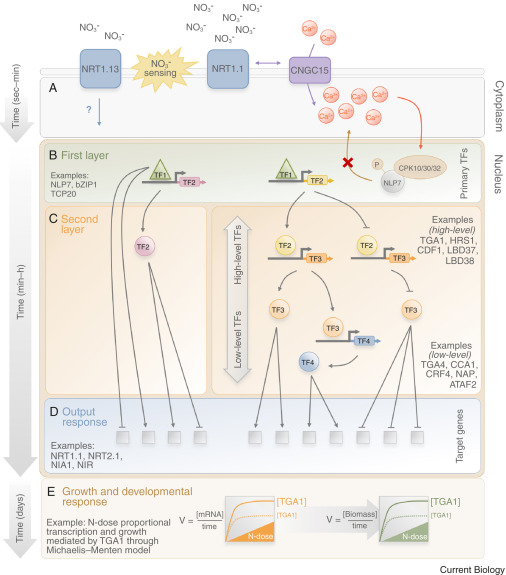
Review: Molecular regulators of nitrate response in plants (Curr. Biol.)
Plant Science Research WeeklyNitrate is the major form of nitrogen used by plants in an aerobic crop cultivation scenario. Lamig et al. review recent additions to the already vast knowledge of nitrate signaling. A first line of regulation concerns nitrate uptake through post-transcriptional and post-translational regulation of nitrate…
ATTED-II v11: A plant gene coexpression database using a sample balancing technique by subagging of principal components (Plant Cell Physiol)
Plant Science Research WeeklyGenes are generally expressed in concert with one another with data often represented by gene co-expression networks. Gene co-expression data may be useful in identifying genes with unknown functions and are generally utilized in exploring molecular processes occurring during normal development as well…
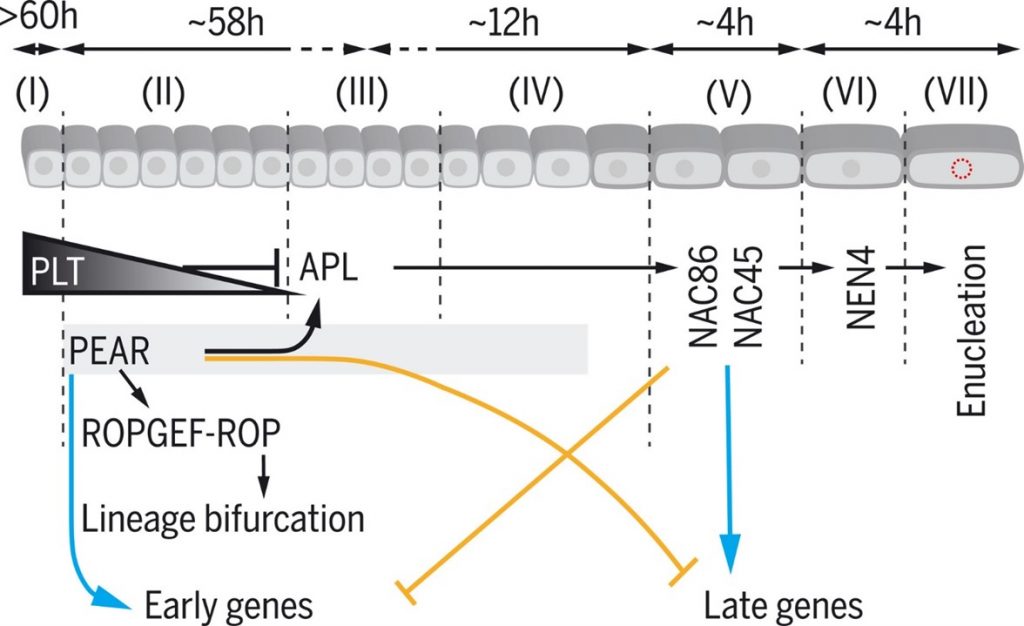
Cell-by-cell dissection of phloem development links a maturation gradient to cell specialization (Science)
Plant Science Research WeeklyIn this seminal paper, Roszak et al. present a deeper understanding of the development of the protophloem cell lineage. The differentiation process from the phloem stem cell to the enucleated cell state spans approximately 19 cells. Using live imaging, FACS sorting and single-cell RNA sequencing, the…
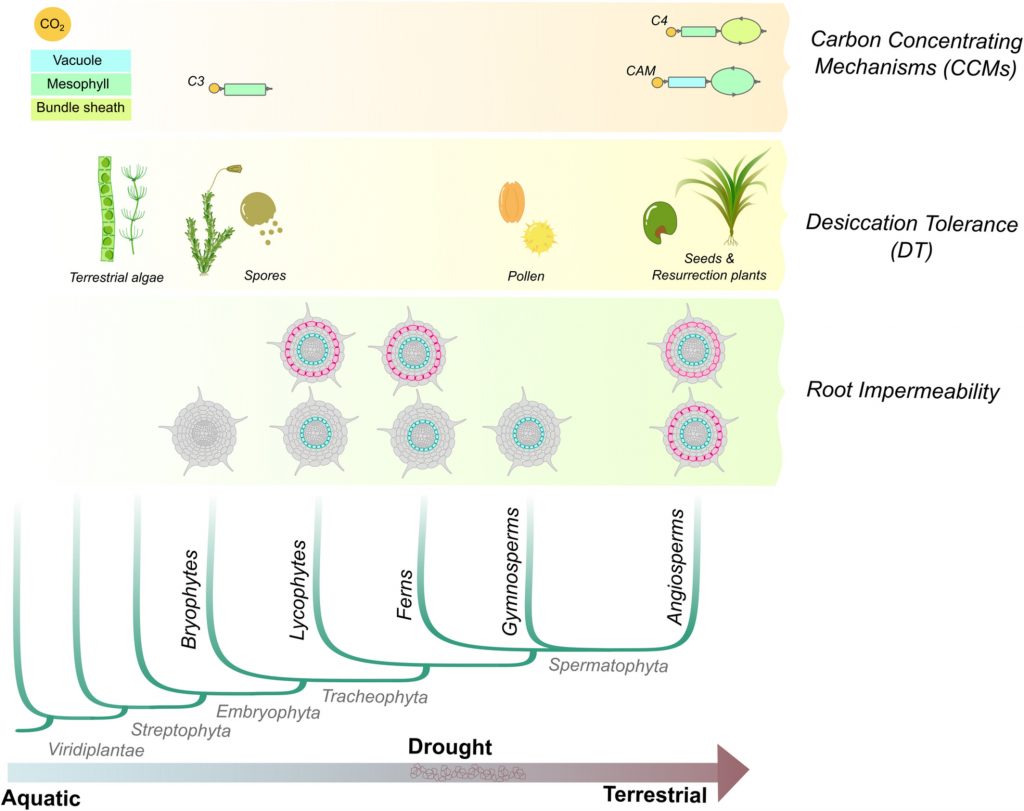
Review: Convergent evolution of gene regulatory networks underlying plant adaptations (Plant Cell Environ.)
Plant Science Research WeeklyThe transition from aquatic to terrestrial habitats exposed plants to low water availability, high light, radiation, and other environmental challenges. To overcome these challenges, plants developed morpho-physiological adaptations to tolerate dry environments and make photosynthesis more efficient.…

ConnecTF: A platform to integrate transcription factor-gene interactions and validate regulatory networks (Plant Physiology)
Plant Science Research WeeklyConnecTF: A platform to integrate transcription factor-gene interactions and validate regulatory networks
Gene regulatory networks (GFNs) are complex beasts, integrating multiple components of transcription factors (TFs), regulatory loci, and function -- not to mention the myriad datasets and methodologies…
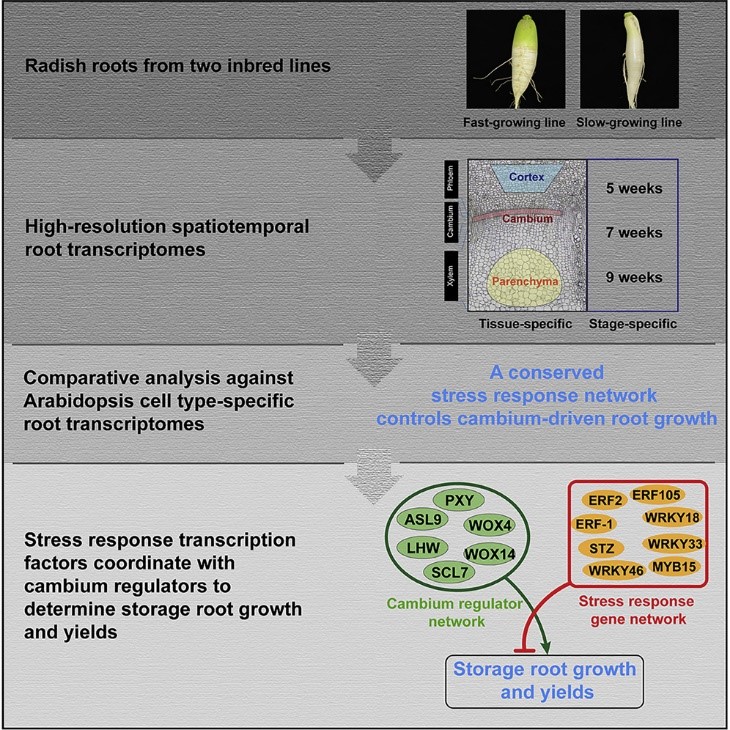
Conserved gene regulatory network integrated with stress response factors in radish and Arabidopsis root cambium (Curr. Biol.)
Plant Science Research WeeklyThe cambium is a layer of actively dividing cells between xylem and phloem tissues that is responsible for the thickening of primary or lateral roots and stems. Root crops are tightly associated with the cambium regulatory mechanism which is less characterized in root as compared to shoot development.…
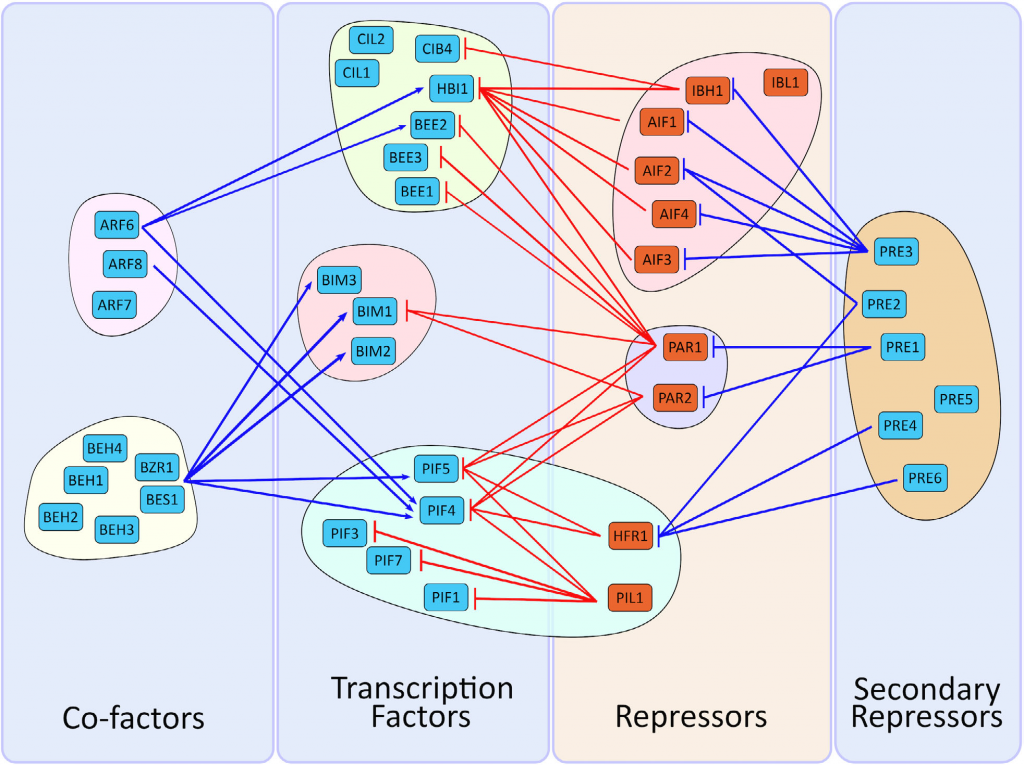
Review: The bHLH network underlying plant shade-avoidance (Physiol. Plant.)
Plant Science Research WeeklyShade avoidance is a complex phenomenon in which plants avoid shade by altering their developmental program in various ways including early flowering, hypocotyl elongation, and more. Many photoreceptors and transcription factors (TFs) are involved in regulating shade avoidance, including the bHLH (basic…

