ConnecTF: A platform to integrate transcription factor-gene interactions and validate regulatory networks (Plant Physiology)
ConnecTF: A platform to integrate transcription factor-gene interactions and validate regulatory networks
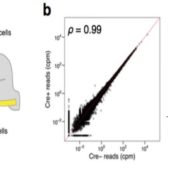
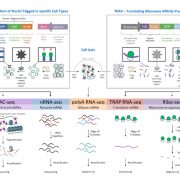
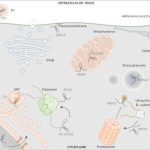
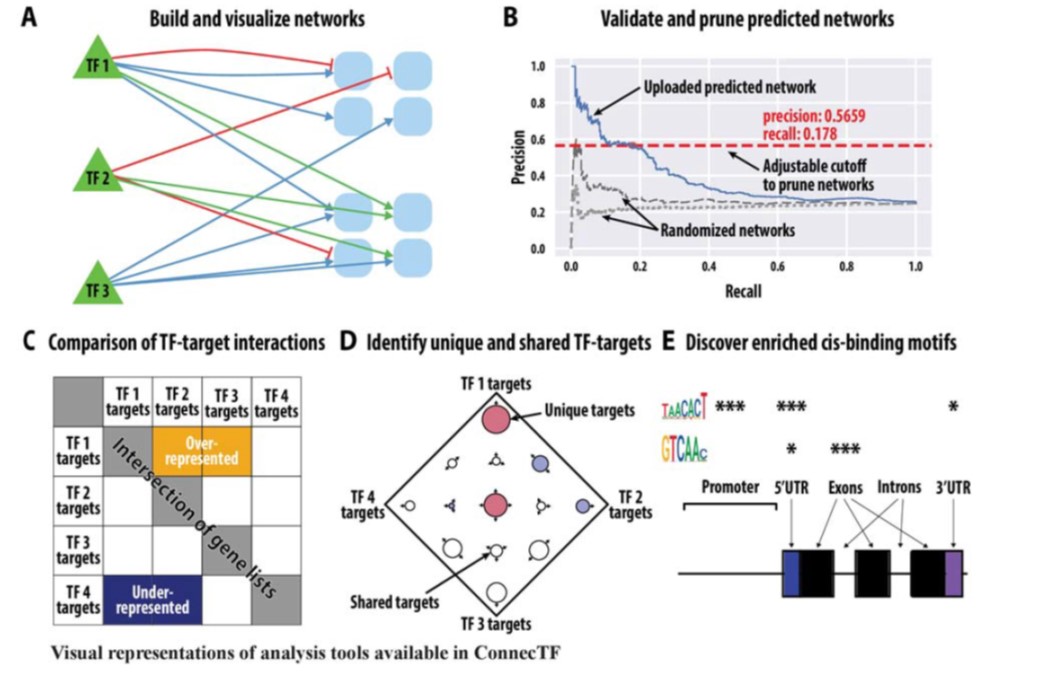 Gene regulatory networks (GFNs) are complex beasts, integrating multiple components of transcription factors (TFs), regulatory loci, and function — not to mention the myriad datasets and methodologies that generate datasets attempting to elucidate such networks. With rapid increases in technology to enable the discovery of more components within these networks, it is necessary to, in parallel, develop methods of organizing such large amounts of data in a user-friendly fashion. Such organizational tools are vital for compiling experimental data together towards the construction of robust GRNs. This was the mission of the authors in this study, who developed and released a web-based, species-independent platform called ConnecTF. ConnecTF, in their words, “integrates genome-wide studies of TF-target binding, TF-target regulation, and other TF-centric omic datasets and uses these to build and refine validated or inferred GRNs.” The authors demonstrate ConnecTF’s power in three case studies: first, identifying potential TF partners for certain TFs involved in ABA signaling; second, refining an inferred GRN for nitrogen signaling; and third, charting a full network path around NLP7, a master TF in nitrogen signaling. Overall, ConnecTF is ripe for use as a user-friendly, publicly available tool to compile and organize data for GRN construction and future GRN-related experiments. Check them out at https://ConnecTF.org! (Summary by Benjamin Jin) Plant Physiology 10.1093/plphys/kiaa012
Gene regulatory networks (GFNs) are complex beasts, integrating multiple components of transcription factors (TFs), regulatory loci, and function — not to mention the myriad datasets and methodologies that generate datasets attempting to elucidate such networks. With rapid increases in technology to enable the discovery of more components within these networks, it is necessary to, in parallel, develop methods of organizing such large amounts of data in a user-friendly fashion. Such organizational tools are vital for compiling experimental data together towards the construction of robust GRNs. This was the mission of the authors in this study, who developed and released a web-based, species-independent platform called ConnecTF. ConnecTF, in their words, “integrates genome-wide studies of TF-target binding, TF-target regulation, and other TF-centric omic datasets and uses these to build and refine validated or inferred GRNs.” The authors demonstrate ConnecTF’s power in three case studies: first, identifying potential TF partners for certain TFs involved in ABA signaling; second, refining an inferred GRN for nitrogen signaling; and third, charting a full network path around NLP7, a master TF in nitrogen signaling. Overall, ConnecTF is ripe for use as a user-friendly, publicly available tool to compile and organize data for GRN construction and future GRN-related experiments. Check them out at https://ConnecTF.org! (Summary by Benjamin Jin) Plant Physiology 10.1093/plphys/kiaa012