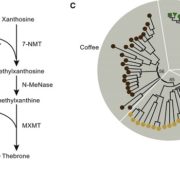
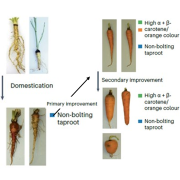
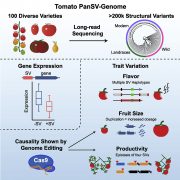
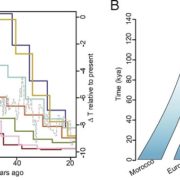
SLAM-ITseq: Sequencing cell type-specific transcriptomes without cell sorting
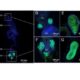
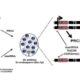
 BioRxiv. Transcriptomic changes at the cellular level are of key importance in specialized cellular types. Therefore, transcriptome analysis at a cell-specific resolution is a powerful tool to learn about biological processes. This analysis is however limited by technological boundaries of microdissection and/or cell sorting methods, such as laser capture microdissection (LCM) or fluorescence-activated cell sorting (FACS). These limitations include intensive time needed, laborious steps and high cell death rate, which limit its successful application to very robust cell-types. Matsushima at al. report a novel sequencing method on living cells that does not make use of previous cell sorting: the SLAM-I(n)T(issue) seq. A new method that improves previously established in vivo 4-thiouracil-based RNA metabolic labelling making it accessible to wider research areas to study cell type-specific transcriptomics, at least in animals. (Summary by Isabel Mendoza) BioRxiv. 10.1101/235093
BioRxiv. Transcriptomic changes at the cellular level are of key importance in specialized cellular types. Therefore, transcriptome analysis at a cell-specific resolution is a powerful tool to learn about biological processes. This analysis is however limited by technological boundaries of microdissection and/or cell sorting methods, such as laser capture microdissection (LCM) or fluorescence-activated cell sorting (FACS). These limitations include intensive time needed, laborious steps and high cell death rate, which limit its successful application to very robust cell-types. Matsushima at al. report a novel sequencing method on living cells that does not make use of previous cell sorting: the SLAM-I(n)T(issue) seq. A new method that improves previously established in vivo 4-thiouracil-based RNA metabolic labelling making it accessible to wider research areas to study cell type-specific transcriptomics, at least in animals. (Summary by Isabel Mendoza) BioRxiv. 10.1101/235093