Transposon-derived small RNAs triggered by miR845 mediate genome dosage response in Arabidopsis
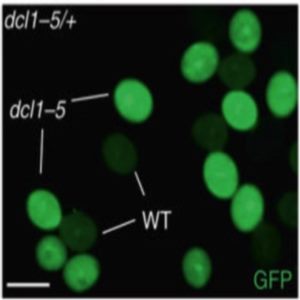 Nat. Genet. Silencing of transposable elements (TEs) is mediated epigenetically by DNA methylation, relying partially on RNA-directed DNA methylation (RdDM). RdDM-induced DNA methylation undergoes a global reprogramming in the male germline, allowing expression of imprinted genes regulating fertility and seed viability. Concomitantly, the production of epigenetically-activated small interfering RNA (easiRNA) from TEs transcripts have been detected, but their biological relevance remains unclear. This study shows that production of TE-derived easiRNA in Arabidopsis pollen is mediated by the targeting of reactivated retrotransposons by microRNA miR485 family. Using a GFP-sensor system including miR485 target site, and thus silenced by easiRNA in pollen, the authors show this phenomenon to be dose-dependent and require RNA polymerase IV, one of the two RNA polymerase involved in RdDM. Remarkably, they show that mutants lacking miR485-dependent easiRNA are less prone to hybridization barrier, between diploid seed parents and tetraploid pollen parents known as triploid block, revealing an important role for easiRNA in regulation of hybridity and speciation. Altogether, this study provides new insights into transposon-derived epigenetic mechanisms to regulation of chromosome dosage and plant development. (Summary by Matthias Benoit) Nat. Genet. 10.1038/s41588-017-0032-5
Nat. Genet. Silencing of transposable elements (TEs) is mediated epigenetically by DNA methylation, relying partially on RNA-directed DNA methylation (RdDM). RdDM-induced DNA methylation undergoes a global reprogramming in the male germline, allowing expression of imprinted genes regulating fertility and seed viability. Concomitantly, the production of epigenetically-activated small interfering RNA (easiRNA) from TEs transcripts have been detected, but their biological relevance remains unclear. This study shows that production of TE-derived easiRNA in Arabidopsis pollen is mediated by the targeting of reactivated retrotransposons by microRNA miR485 family. Using a GFP-sensor system including miR485 target site, and thus silenced by easiRNA in pollen, the authors show this phenomenon to be dose-dependent and require RNA polymerase IV, one of the two RNA polymerase involved in RdDM. Remarkably, they show that mutants lacking miR485-dependent easiRNA are less prone to hybridization barrier, between diploid seed parents and tetraploid pollen parents known as triploid block, revealing an important role for easiRNA in regulation of hybridity and speciation. Altogether, this study provides new insights into transposon-derived epigenetic mechanisms to regulation of chromosome dosage and plant development. (Summary by Matthias Benoit) Nat. Genet. 10.1038/s41588-017-0032-5