Translational regulation of PHOSPHATE 1 affects shoot phosphate levels and biomass
Eva Hellmann, The Sainsbury Laboratory, University of Cambridge, CB2 1LR Cambridge, United Kingdom
Phosphorus (P) is an essential plant nutrient and makes up part of membranes, DNA and RNA; it also serves as energy currency in the form of ATP cycling. It is involved in many signaling processes enabling proper growth and development. The availability of phosphorus in the soil is often limited, and to mobilize it, plants have developed a range of transporters. Agriculture relies heavily on the usage of phosphate fertilizers that are produced from phosphorus-rich rocks, a finite resource.
The main exporter that transports phosphate into the xylem is PHOSPHATE 1 (PHO1) (Hamburger et al., 2002). Loss of PHO1 leads to stunted growth and low shoot phosphate levels (Rouached et al., 2011). PHO1 was shown to be regulated mainly at the post-transcriptional level by ubiquitination (Liu et al., 2012) or by a natural antisense RNA (Jabnoune et al., 2013).
Genes that are regulated at the translational level often contain small upstream reading frames (uORFs) in front of their main reading frame (mORF). In Arabidopsis thaliana, this is true for about one third of the transcriptome (Srivastava et al., 2018). uORFs enhance or suppress mORF translation dependent on context, conservation, distance, number and overlap with each other and the mORF (Calvo et al., 2009).
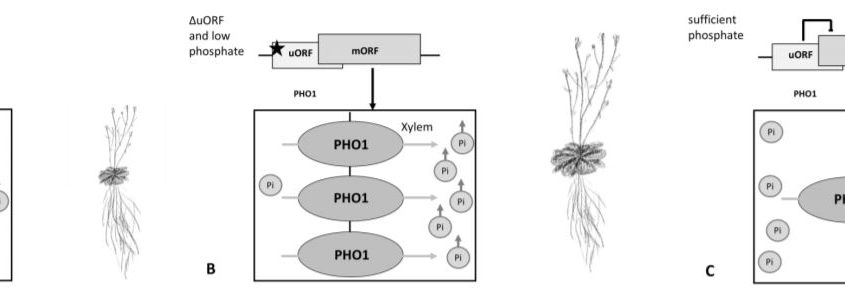
In their recent study, Reis et al (2020) unravel the function and regulation of PHO1, focusing on its translational regulation via an uORF. The authors identified uORFs upstream of the PHO1 gene in various plant species. In Arabidopsis and maize (Zea mays), the uORF is short and overlaps with the start of the mORF. This is often the case for uORFs that repress the translation of the mORF. By analyzing previously published data, the authors found that the PHO1 transcript is generally poorly associated with ribosomes, indicating a low translational efficiency which is consistent with a possible inhibitory effect of the uORF (Deforges et al., 2019). The authors showed evidence for this by mutating the uORF start codon (∆uORF), which in protoplast luciferase assays led to a 20-fold increase in luminescence.
The authors also found that mutation of the uORF leads to higher association of the transcript with polysomes, suggesting a higher rate of translation, whilst transcript levels were unaffected. Furthermore, in plants harboring the mutated uORF, PHO1 protein levels are higher than in the wild-type plants, based on immunoblotting, in sufficient and low phosphate conditions.
To analyze the role of the uORF in growth and shoot phosphate accumulation, the authors examined pho1 mutants that were transformed with PHO1 complementation constructs containing the wild-type uORF or ∆uORF. Under sufficient or high phosphorus conditions, the pho1 mutant shoot phosphate levels and growth phenotype were complemented with both wild type uORF or ∆uORF constructs. In low phosphate conditions however, plants transformed with ∆uORF exhibited twice the shoot mass and higher shoot phosphate levels than plants transformed with the wild-type uORF.
Next, the authors identified natural Arabidopsis accessions with disruptive short nucleotide polymorphisms (SNPs) in the PHO1 uORF. By analyzing the F2 progeny of an accession with mutated uORF crossed with Col-0, the authors showed that high shoot phosphate levels segregate with a mutated uORF.
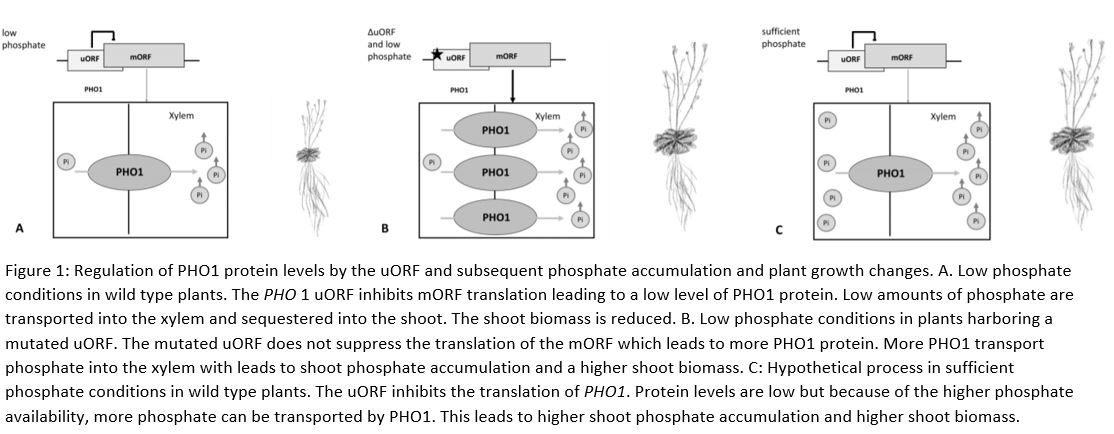
This study beautifully unveils how the PHO1 uORF regulates PHO1 translationUnder low phosphate conditions, the PHO1 uORF inhibits the readout of the mORF, leading to less PHO1 protein, less phosphate transport into the xylem, less shoot phosphate accumulation and decreased growth. If the uORF is lost, there is more PHO1 protein, and even in depleted soil more transport of phosphate into the xylem leading to more shootward accumulation and enhanced growth (Fig 1 A, B).
One could speculate, that in sufficient phosphate conditions, the uORF might inhibit PHO1 translation but the phosphate levels would still enable vigorous shoot growth and shoot phosphate accumulation (Fig 1 C). It would be interesting to see if protein levels or the inhibition of the mORF by the uORF are phosphate-dependent and if so, how this is realized.
The absence of the PHO1 uORF in several natural Arabidopsis accessions suggests that there is no strong selective pressure to regulate PHO1 levels through the uORF. Therefore, it might be feasible to remove the PHO1 uORF in crops to increase PHO1 protein levels and improve crop yields on low phosphate soils independently of fertilizer application. Given the similarity of the Arabidopsis and maize PHO1 uORF, this would be an exciting and promising undertaking that we would love to read more about in the future.
Literature Cited
Calvo SE, Pagliarini DJ, Mootha VK (2009) Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci U S A 106: 7507–7512
Deforges J, Reis RS, Jacquet P, Sheppard S, Gadekar VP, Hart-Smith G, Tanzer A, Hofacker IL, Iseli C, Xenarios I, et al (2019) Control of Cognate Sense mRNA Translation by cis-Natural Antisense RNAs. Plant Physiol 180: 305–322
Hamburger D, Rezzonico E, MacDonald-Comber Petétot J, Somerville C, Poirier Y (2002) Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 14: 889–902
Jabnoune M, Secco D, Lecampion C, Robaglia C, Shu Q, Poirier Y (2013) A Rice cis-Natural antisense RNA acts as a translational enhancer for its cognate mRNA and contributes to phosphate homeostasis and plant fitness. Plant Cell 25: 4166–4182
Liu TY, Huang TK, Tseng CY, Lai YS, Lin SI, Lin WY, Chen JW, Chioua TJ (2012) PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell 24: 2168–2183
Rouached H, Stefanovic A, Secco D, Bulak Arpat A, Gout E, Bligny R, Poirier Y (2011) Uncoupling phosphate deficiency from its major effects on growth and transcriptome via PHO1 expression in Arabidopsis. Plant J 65: 557–570
Srivastava AK, Lu Y, Zinta G, Lang Z, Zhu J-K (2018) UTR-Dependent Control of Gene Expression in Plants. Trends Plant Sci 23: 248–259