Ticket to Ride: tRNA-Related Sequences and Systemic Movement of mRNAs
IN BRIEF by Jennifer Mach [email protected]
Movement of macromolecules through the plant phloem provides a mechanism for long-distance signaling that plants use in development, disease resistance, and other adaptive responses (reviewed in Spiegelman et al., 2013). For example, full-length RNAs, such as the Arabidopsis thaliana GIBBERELLIC ACID INSENSITIVE and potato (Solanum tuberosum) BEL5 mRNAs, and small RNAs, including numerous microRNAs involved in gene silencing, move through the phloem to affect distal regions of the plant. Emerging evidence indicates that RNA transport occurs more pervasively than indicated by the isolated examples first identified. Indeed, a recent study used grafted plants of different ecotypes to identify 2006 Arabidopsis mRNAs that moved across the graft junction (Thieme et al., 2015).
What causes RNAs to move? Recent work (Calderwood et al., 2016; highlighted in Hofmann, 2016) used modeling to show that mRNA abundance in the phloem companion cell can explain mRNA mobility; simply, the more abundant the mRNA in these cells, the more likely it is to end up in phloem sap. The abundance model does not preclude selective transport and does not explain the mobility (and nonmobility) of some mRNAs. Therefore, polypyrimidine tract sequences or tRNA-like structures (TLSs) might provide a mechanistic and evolutionary explanation for the mobility of some specific mRNAs.
TLSs have a tRNA-like stem-loop secondary structure and play a key role in the infectivity of RNA viruses. To test the role of TLSs in the mobility of plant transcripts, Zhang et al. (2016) used DISRUPTION OF MEIOTIC CONTROL1 (DMC1); silencing of DMC1, or the presence of a truncated, dominant-negative DMC1 (DNDMC1) causes pollen defects and male sterility. Thus, male sterility provides a readout for translocation of DNDMC1 mRNA fusions moving from the rootstock into a grafted, wild-type scion. The DNDMC1 RNA sequences do not contain mobility motifs; using this transcript, the authors tested several constructs in transgenic tobacco (Nicotiana tabacum), including DNDMC1 fused to the mobile transcript of potato BEL5 and the nonmobile transcript of tobacco CENTRORADIALIS-like 2. As expected, plants expressing DNDMC1 showed male sterility, and wild-type scions grafted onto rootstocks producing DNDMC1–BEL5 mRNA showed high percentages of aberrant pollen. In addition to showing that this fusion can translocate, this observation also shows that this translocated mRNA can be translated at its destination.
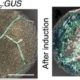
 Using this reporter system, the authors next tested whether tRNA sequences can trigger movement of the DNDMC1 mRNA. tRNAMet occurs in phloem sap, so the authors also made tRNAMet–DNDMC1 fusion constructs and transgenic plants. Fusion of tRNAMet to the 5′ or 3′ untranslated region allowed the DNDMC1 mRNA to move across the graft junction, causing male sterility in the wild-type scion. The authors tested for potential induction of systemic silencing, using a YFP-DNDMC1 line, but found strong YFP signals, indicating that the mobile tRNA fusion construct did not produce small interfering RNAs that induced silencing. The authors also tested tRNA sequences in Arabidopsis, using GUS fusions to tRNAMet and tRNAGly, which are imported into the phloem, and tRNAIle, which is not imported into the phloem. Staining for GUS activity showed that the phloem-imported tRNAs could mediate transport of tRNA-GUS mRNA from root to shoot and from shoot to root. By contrast, grafting to GUS-tRNAIle or 35S:GUS plants did not result in GUS activity in the grafted wild-type tissue. Deletion analysis of tRNAMet showed that removal of the anticodon and TψC hairpin-loop abolished mRNA mobility.
Using this reporter system, the authors next tested whether tRNA sequences can trigger movement of the DNDMC1 mRNA. tRNAMet occurs in phloem sap, so the authors also made tRNAMet–DNDMC1 fusion constructs and transgenic plants. Fusion of tRNAMet to the 5′ or 3′ untranslated region allowed the DNDMC1 mRNA to move across the graft junction, causing male sterility in the wild-type scion. The authors tested for potential induction of systemic silencing, using a YFP-DNDMC1 line, but found strong YFP signals, indicating that the mobile tRNA fusion construct did not produce small interfering RNAs that induced silencing. The authors also tested tRNA sequences in Arabidopsis, using GUS fusions to tRNAMet and tRNAGly, which are imported into the phloem, and tRNAIle, which is not imported into the phloem. Staining for GUS activity showed that the phloem-imported tRNAs could mediate transport of tRNA-GUS mRNA from root to shoot and from shoot to root. By contrast, grafting to GUS-tRNAIle or 35S:GUS plants did not result in GUS activity in the grafted wild-type tissue. Deletion analysis of tRNAMet showed that removal of the anticodon and TψC hairpin-loop abolished mRNA mobility.
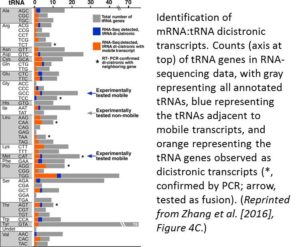
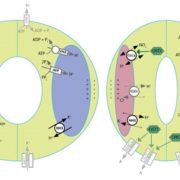
Finding that the tRNA sequences could induce RNA mobility raised the question of whether this occurred for endogenous RNAs. Database analysis and secondary structure predictions indicated that known Arabidopsis mobile transcripts are enriched in sequences that could form a TLS. Also, tRNA genes tend to be closer in the genome to loci encoding mobile mRNAs than to genes encoding nonmobile mRNAs. Analysis of RNA-sequencing data indicated that these nearby genes can form dicistronic mRNA-tRNA transcripts at a high frequency (see figure). For example, CHOLINE KINASE1 (CK1) produces a mobile transcript and the RNA-sequencing data included a CK1-tRNAGly dicistronic transcript. Indeed, in a T-DNA insertion mutant that separated CK1 from tRNAGly, the CK1 transcript failed to move across graft junctions.
Thus, this emerging research shows that in addition to potential abundance-based movement into the phloem, some RNAs grab a tRNA ticket for a ride through the phloem and that the joined mRNA can be translated after transport. Such active transport, which can occur bidirectionally, and the specific mechanisms for routing these RNAs to their destination, remain intriguing questions for the future.
REFERENCES
Calderwood, A., Kopriva, S., Morris, R.J. (2016). Transcript abundance explains mRNA mobility data in Arabidopsis thaliana. Plant Cell 28: 610–615.
Hofmann, N.R. (2016). Should I stay or should I go? Abundance as a new null hypothesis for determination of mRNA mobility. Plant Cell 28: 597–598.
Spiegelman, Z., Golan, G., Wolf, S. (2013). Don’t kill the messenger: Long-distance trafficking of mRNA molecules. Plant Sci. 213: 1–8.
Thieme, C.J., Rojas-Triana, M., Stecyk, E., Schudoma, C., Zhang, W., Yang, L., Miñambres, M., Walther, D., Schulze, W.X., Paz-Ares, J., Scheible, W.-R., Kragler, F.(2015). Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nat. Plants 1: 15025.
Zhang, W., Thieme, C., Kollwig, G., Apelt, F., Yang, L., Winter, W., Andresen, N., Walther, D., Kragler, F. (2016). tRNA-related sequences trigger systemic mRNA transport in plants. Plant Cell 28: 1237–1249.





Leave a Reply
Want to join the discussion?Feel free to contribute!