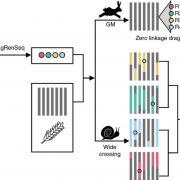
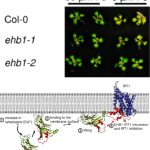
NRT1.1B is associated with root microbiota composition and nitrogen use in rice ($) (Nature Biotechnol)
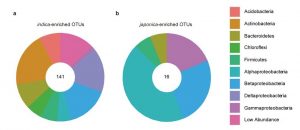 Availability of nitrogen has always been a limiting factor for rice production. Different soil inhabitants and root-associated bacterial populations are involved in making nitrogen available to plants in organic as well as in inorganic forms. However, plant absorption of nitrogen is selective and affects its nitrogen use efficiency (NUE). The two main Asian cultivated rice varieties, japonica and indica have different NUE, attributed in part to the natural variation in a gene called NRT1.1B that encodes a nitrate transporter and sensor. By using 68 indica and 27 japonica varieties, Zhang et al. found a relationship between NRT1.1B and rice root-associated microbial populations. The microbiome community structure analysis from both varieties showed a clear influence of genotype effect, moreover the indica variety showed higher microbial diversity than japonica. Functional assignment to the microbial population showed more of the indica associated microbes are involved in nitrogen metabolism function. The authors also generated a synthetic microbial community based on genotype-specific and NRT1.1B-associated observations by using established culture collections. (Summary by Mugdha Sabale) Nature Biotechnol. 10.1038/s41587-019-0104-4
Availability of nitrogen has always been a limiting factor for rice production. Different soil inhabitants and root-associated bacterial populations are involved in making nitrogen available to plants in organic as well as in inorganic forms. However, plant absorption of nitrogen is selective and affects its nitrogen use efficiency (NUE). The two main Asian cultivated rice varieties, japonica and indica have different NUE, attributed in part to the natural variation in a gene called NRT1.1B that encodes a nitrate transporter and sensor. By using 68 indica and 27 japonica varieties, Zhang et al. found a relationship between NRT1.1B and rice root-associated microbial populations. The microbiome community structure analysis from both varieties showed a clear influence of genotype effect, moreover the indica variety showed higher microbial diversity than japonica. Functional assignment to the microbial population showed more of the indica associated microbes are involved in nitrogen metabolism function. The authors also generated a synthetic microbial community based on genotype-specific and NRT1.1B-associated observations by using established culture collections. (Summary by Mugdha Sabale) Nature Biotechnol. 10.1038/s41587-019-0104-4