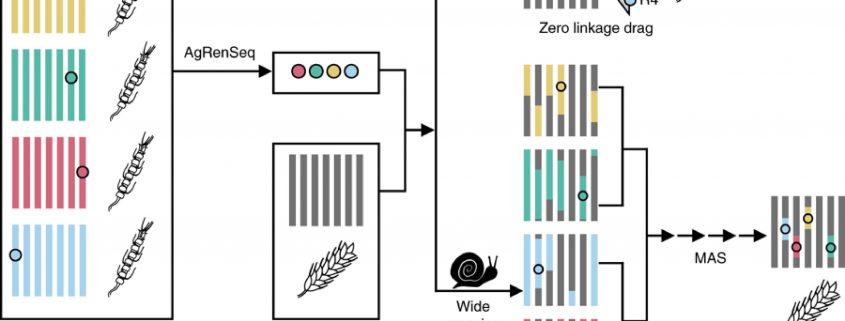
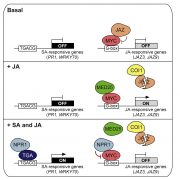
Tapping into the genetic diversity of wild crops for engineering disease resistance (Nature Biotech)
Lack of sequence information makes harnessing the diversity of disease resistance (R) genes in wild crops highly challenging. Arora and Steuernagel et al. report AgRenSeq, a reference genome-free technique, for rapid cloning of nucleotide binding/Leucine-rich repeat (NLR) resistance genes from wild crops. Using R gene enrichment sequencing, the authors captured thousands of NLR-specific contigs from 151 distinct accessions of Aegilops taushii, a wild relative of bread wheat. Based on k-mer based association mapping, they identified four wheat stem rust resistance genes from the captured contigs. AgRenSeq thus provides a powerful technique for discovering rare, uncharacterized R genes from genetically diverse accessions of any crop and improving disease resistance in its domesticated counterparts. (Summary by Saima Shahid) Nature Biotech. 10.1038/s41587-018-0007-9.