Snapshot of a TF Network in Plants
Li et al. investigate a transcription factor network that transmits environmental signals to regulate secondary metabolism in plants. The Plant Cell https://doi.org/10.1105/tpc.17.00805.
By Baohua Li and Daniel Kliebenstein
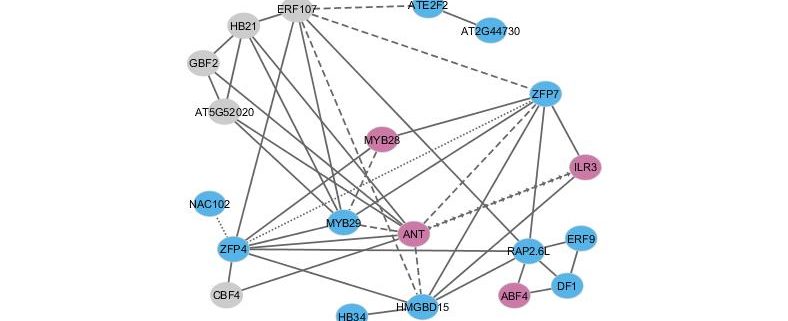
Background: Plants produce specialized secondary metabolites to survive biotic and abiotic stress. Secondary metabolite accumulation in plants is strongly influenced by transcription factors (TFs), which play key roles in regulating target gene expression. TFs also play crucial roles in communicating environmental changes, which lead to changes in the accumulation of secondary metabolites. While the effects and functions of some single plant secondary metabolite-related TFs in a fixed environment have been reported, little known about how these TFs interact with each other under different environmental conditions to control secondary metabolite accumulation. We used a model plant secondary metabolic pathway, the glucosinolates (GLS) pathway, in Arabidopsis to conduct a systematic survey to explore the principles underlying how an organism’s genes work together to transmit information about environmental fluctuations.
Question: We tested how TFs genetically interact with each other under different environmental conditions to influence the accumulation of GLS. We did this by systematically designing, producing, and studying GLS accumulation in loss-of-function mutants of two or three TFs in Arabidopsis under different environmental conditions.
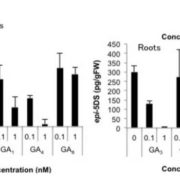
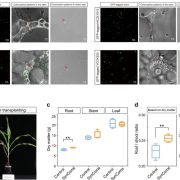
Findings: There are widespread epistatic interactions between TFs that regulate GLS accumulation. The modes and effects of these TF-TF interactions change depending on the GLS phenotypes. Short-chain aliphatic GLS are the major targets for the networks under analysis. The combined effects of the TFs on these GLS are generally smaller than what we would predict from the analysis of single mutants. Environmental conditions play complex roles in the effects of TF networks on GLS accumulation. By combining data from expression analysis of individual GLS pathway genes, the activities of key enzymes of the GLS pathway, and final GLS accumulation, we developed a model for how the TF network shifts and changes dynamically to control the GLS pathway at these three different levels.
Next steps: Although we performed one of the largest plant metabolism studies to date, it is still like taking a cup of water from the Mississippi River. We are currently building a larger TF network, and we are interested in studying how the same TF network coordinates plant metabolism with plant development.
Li, B., Tang, M., Nelson, A., Caligagan, H., Zhou, X., Clark-Wiest, C., Ngo, R., Brady, S.M., and Kliebenstein, D.J. (2018). Network-guided Discovery of Extensive Epistasis Between Transcription Factors Involved in Aliphatic Glucosinolate Biosynthesis. https://doi.org/10.1105/tpc.17.00805.