Signatures of adaptation to mutualists revealed by root transcriptional dynamics
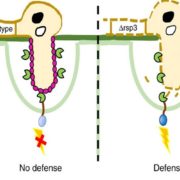
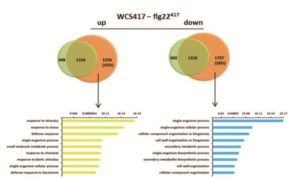 A plant’s rhizosphere consists of a huge array of pathogenic microbes many of which can trigger defense responses, leading to decreased growth. On the other hand, beneficial microbes such as rhizobacteria promote growth and can induce systemic resistance while suppressing local immune responses. A recent study aimed to understand the differential host responses towards the colonizing pathogenic or beneficial rhizobacteria like Pseudomonas simiae (WCS417). Stringlis et al. carried out a comprehensive time-resolved RNA-seq of Arabidopsis roots in response to either the rhizobacteria WCS417 strain or the pathogenic epitope MAMPs flg22417 (from WCS417), flg22Pa (from pathogenic P. aeruginosa) or fungal chitin. The transcriptional responses display a large overlap of 74% of genes between different treatments, although they differ temporally. WCS417 was unique in its ability to actively suppress MAMP-triggered transcriptional responses. Additional comparison of flg22417 to WCS417 transcriptional responses demonstrated a high proportion of auxin-related genes regulated during rhizobia colonization. This study provides evidence for a dual role of auxin signaling in balancing growth and defense responses upon colonization of plants by growth promoting rhizobacteria. (Summary by Amey Redkar) Plant J. 10.1111/tpj.13741/abstract
A plant’s rhizosphere consists of a huge array of pathogenic microbes many of which can trigger defense responses, leading to decreased growth. On the other hand, beneficial microbes such as rhizobacteria promote growth and can induce systemic resistance while suppressing local immune responses. A recent study aimed to understand the differential host responses towards the colonizing pathogenic or beneficial rhizobacteria like Pseudomonas simiae (WCS417). Stringlis et al. carried out a comprehensive time-resolved RNA-seq of Arabidopsis roots in response to either the rhizobacteria WCS417 strain or the pathogenic epitope MAMPs flg22417 (from WCS417), flg22Pa (from pathogenic P. aeruginosa) or fungal chitin. The transcriptional responses display a large overlap of 74% of genes between different treatments, although they differ temporally. WCS417 was unique in its ability to actively suppress MAMP-triggered transcriptional responses. Additional comparison of flg22417 to WCS417 transcriptional responses demonstrated a high proportion of auxin-related genes regulated during rhizobia colonization. This study provides evidence for a dual role of auxin signaling in balancing growth and defense responses upon colonization of plants by growth promoting rhizobacteria. (Summary by Amey Redkar) Plant J. 10.1111/tpj.13741/abstract