Phloem unloading in Arabidopsis roots
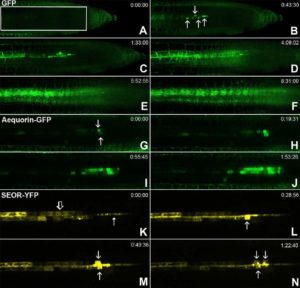 It is well known that long distance transport and movement of molecules is enabled by phloem, but the precise mechanism of loading/unloading of phloem mobile compounds is not known. In this article, Ross-Elliott et al. used a combination of approaches (non-invasive imaging, 3D-electron microscopy, and mathematical modelling) to explore phloem unloading. The authors show that unloading of solutes in Arabidopsis roots occurs through plasmodesmata by convective unloading, a combination of mass flow and diffusion. By contrast, unloading of small proteins occurs in pulses, which they describe as batch unloading, through specialized funnel plasmodesmata, into the phloem pole pericycle. The authors also propose a new model of phloem unloading in Arabidopsis roots. (Summary by Mather Khan) eLife 10.7554/eLife.24125
It is well known that long distance transport and movement of molecules is enabled by phloem, but the precise mechanism of loading/unloading of phloem mobile compounds is not known. In this article, Ross-Elliott et al. used a combination of approaches (non-invasive imaging, 3D-electron microscopy, and mathematical modelling) to explore phloem unloading. The authors show that unloading of solutes in Arabidopsis roots occurs through plasmodesmata by convective unloading, a combination of mass flow and diffusion. By contrast, unloading of small proteins occurs in pulses, which they describe as batch unloading, through specialized funnel plasmodesmata, into the phloem pole pericycle. The authors also propose a new model of phloem unloading in Arabidopsis roots. (Summary by Mather Khan) eLife 10.7554/eLife.24125



 This resource provides a set of videos and a practical investigation aimed at supporting working scientifically in the classroom and relating science to real world experiences. In the first video Professor Brian Cox joins a teacher to find out how to set up and run an investigation to find out if plants need soil to grow. Children try to germinate and grow plants from a seed using a variety of different materials instead of soil. Further videos show Brian Cox visiting an Industrial farm to find out about how they grow vegetables in a building and meeting a researcher looking at soil health.
This resource provides a set of videos and a practical investigation aimed at supporting working scientifically in the classroom and relating science to real world experiences. In the first video Professor Brian Cox joins a teacher to find out how to set up and run an investigation to find out if plants need soil to grow. Children try to germinate and grow plants from a seed using a variety of different materials instead of soil. Further videos show Brian Cox visiting an Industrial farm to find out about how they grow vegetables in a building and meeting a researcher looking at soil health. If you’re applying for an independent position, you’ll need to submit a Research Statement about your future research plans. Bethany Hout (
If you’re applying for an independent position, you’ll need to submit a Research Statement about your future research plans. Bethany Hout ( Some bees are generalist pollinators that gather pollen from a wide range of species, whereas others are specialists that visit only one or a few species. Vanderplanck et al. examined floral traits of the host plants of two different groups of generalist bees. There was no significant correlation between bees and the floral scent, floral reflectance, and total amino acid composition in pollen of the flowers they visit. By contrast, pollen sterol content and polypeptide content does correlate with bee preferences. This study provides insights into the evolutionary forces that affect host-shifts in bees. Sci. Reports
Some bees are generalist pollinators that gather pollen from a wide range of species, whereas others are specialists that visit only one or a few species. Vanderplanck et al. examined floral traits of the host plants of two different groups of generalist bees. There was no significant correlation between bees and the floral scent, floral reflectance, and total amino acid composition in pollen of the flowers they visit. By contrast, pollen sterol content and polypeptide content does correlate with bee preferences. This study provides insights into the evolutionary forces that affect host-shifts in bees. Sci. Reports 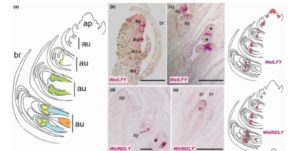 In order to look at the origins of flowers, Moyroud et al. looked at reproductive controls in the gymnosperm Welwitschia mirabilis, which, as the authors say, is a good model because, “Although the plant body is famously bizarre, the reproductive structures are generalized.” Furthermore, its male reproductive structures are produced as soon as two years after germination, facilitating study. The authors found orthologues of key flowering genes in Welwitschia, and showed that, like its angiosperm counterpart, Welwitschia LFY binds to the promoter of MADS box genes corresponding to AP3/PI. Furthermore, they found that early in their development male cones showed B- and C-class MADS-box gene expression, whereas female cones showed C class expression; the same pattern as observed in angiosperms. Therefore, Welwitschia mirabilis sheds light on ancestral mechanisms prefiguring floral development. New Phytol.
In order to look at the origins of flowers, Moyroud et al. looked at reproductive controls in the gymnosperm Welwitschia mirabilis, which, as the authors say, is a good model because, “Although the plant body is famously bizarre, the reproductive structures are generalized.” Furthermore, its male reproductive structures are produced as soon as two years after germination, facilitating study. The authors found orthologues of key flowering genes in Welwitschia, and showed that, like its angiosperm counterpart, Welwitschia LFY binds to the promoter of MADS box genes corresponding to AP3/PI. Furthermore, they found that early in their development male cones showed B- and C-class MADS-box gene expression, whereas female cones showed C class expression; the same pattern as observed in angiosperms. Therefore, Welwitschia mirabilis sheds light on ancestral mechanisms prefiguring floral development. New Phytol. 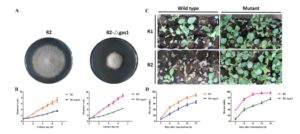 Fusarium oxysporum is a fungal pathogen of plants. F. oxysporum f. sp. conglutinans (Foc) causes fungal wilt in cabbage. Two races have been identified, with Race 2 being much more pathogenic than Race 1. Li et al. used a proteomic approach to investigate the origin of Race 2’s enhanced pathogenicity. They observed 145 proteins that differ in abundance between the two races. Proteins involved in inorganic ion transport and carbohydrate transport and metabolism were notably more abundant in the more pathogenic race, as were their corresponding mRNAs. Knocking out a gene in encoding one of these differentially expressed proteins (a glucanosyltransferase) significantly lowered the pathogenicity of both races. Sci. Reports
Fusarium oxysporum is a fungal pathogen of plants. F. oxysporum f. sp. conglutinans (Foc) causes fungal wilt in cabbage. Two races have been identified, with Race 2 being much more pathogenic than Race 1. Li et al. used a proteomic approach to investigate the origin of Race 2’s enhanced pathogenicity. They observed 145 proteins that differ in abundance between the two races. Proteins involved in inorganic ion transport and carbohydrate transport and metabolism were notably more abundant in the more pathogenic race, as were their corresponding mRNAs. Knocking out a gene in encoding one of these differentially expressed proteins (a glucanosyltransferase) significantly lowered the pathogenicity of both races. Sci. Reports 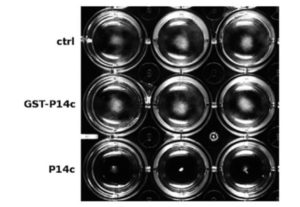 PATHOGENESIS-RELATED 1 (PR-1) protein was identified 50 years ago as a small protein induced in response to pathogens, but its mode of action has remained obscure. PR-1 is a member of the CAP family (cysteine-rich secretory protein, antigen 5, and pathogenesis-related 1). These proteins share a 150 amino acid domain that forms an α-β-α sandwich fold. Recently, yeast CAP proteins were found to bind sterols. Gamir et al. showed that PR-1 also can bind sterols, and that expression of PR-1 can complement yeast CAP mutants. Phytophthora are unable to synthesize sterols and rely on their uptake from the medium; these oomycetes are also much more sensitive to PR-1 than fungal pathogens. The authors showed that PR-1 can inhibit growth of Phytophthora brassicae in vitro, but that this inhibition is eliminated in the presence of excess cholesterol in the growth medium. PR-1 mutants blocked in sterol binding, or fusion proteins rendered too large to pass into the pathogen cell lack antimicrobial activity. Together, these studies suggest that PR-1’s mode of action is through direct removal of sterols from pathogens. Plant J.
PATHOGENESIS-RELATED 1 (PR-1) protein was identified 50 years ago as a small protein induced in response to pathogens, but its mode of action has remained obscure. PR-1 is a member of the CAP family (cysteine-rich secretory protein, antigen 5, and pathogenesis-related 1). These proteins share a 150 amino acid domain that forms an α-β-α sandwich fold. Recently, yeast CAP proteins were found to bind sterols. Gamir et al. showed that PR-1 also can bind sterols, and that expression of PR-1 can complement yeast CAP mutants. Phytophthora are unable to synthesize sterols and rely on their uptake from the medium; these oomycetes are also much more sensitive to PR-1 than fungal pathogens. The authors showed that PR-1 can inhibit growth of Phytophthora brassicae in vitro, but that this inhibition is eliminated in the presence of excess cholesterol in the growth medium. PR-1 mutants blocked in sterol binding, or fusion proteins rendered too large to pass into the pathogen cell lack antimicrobial activity. Together, these studies suggest that PR-1’s mode of action is through direct removal of sterols from pathogens. Plant J. 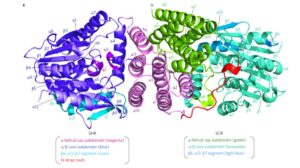 Structural biology provides a key link between genotype and phenotype. Hirano et al. probe the structure of a heterodimer of plant-specific GRAS family transcriptional regulators [SHORT-ROOT (SHR) and SCARECROW (SCR)] as bound to the transcription factor JACKDAW (JKD). The GRAS family proteins (encoded by 33 genes in Arabidopsis and 66 in rice) are involved in several signaling pathways, including root development and gibberellin and phytochrome signaling. Their analysis suggests that in GRAS proteins, the alpha-helical cap domain affects whether the proteins homo- or hetero-dimerize. The authors also found no evidence for direct DNA binding of the SHR-SCR complex, and propose that it acts as a transcription cofactors that requires complex formation with JKD for DNA binding. Nature Plants
Structural biology provides a key link between genotype and phenotype. Hirano et al. probe the structure of a heterodimer of plant-specific GRAS family transcriptional regulators [SHORT-ROOT (SHR) and SCARECROW (SCR)] as bound to the transcription factor JACKDAW (JKD). The GRAS family proteins (encoded by 33 genes in Arabidopsis and 66 in rice) are involved in several signaling pathways, including root development and gibberellin and phytochrome signaling. Their analysis suggests that in GRAS proteins, the alpha-helical cap domain affects whether the proteins homo- or hetero-dimerize. The authors also found no evidence for direct DNA binding of the SHR-SCR complex, and propose that it acts as a transcription cofactors that requires complex formation with JKD for DNA binding. Nature Plants