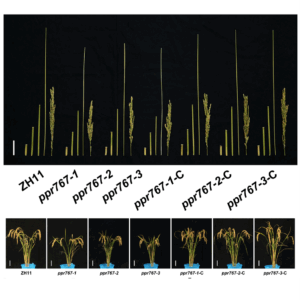
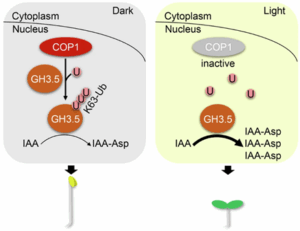
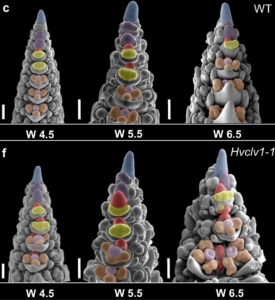
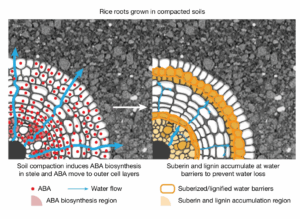
Review: Lost in translation – lessons from concepts that don’t translate from Arabidopsis
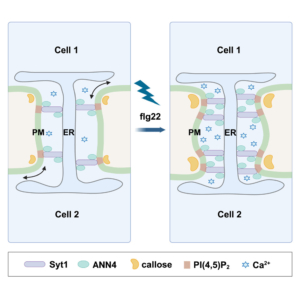
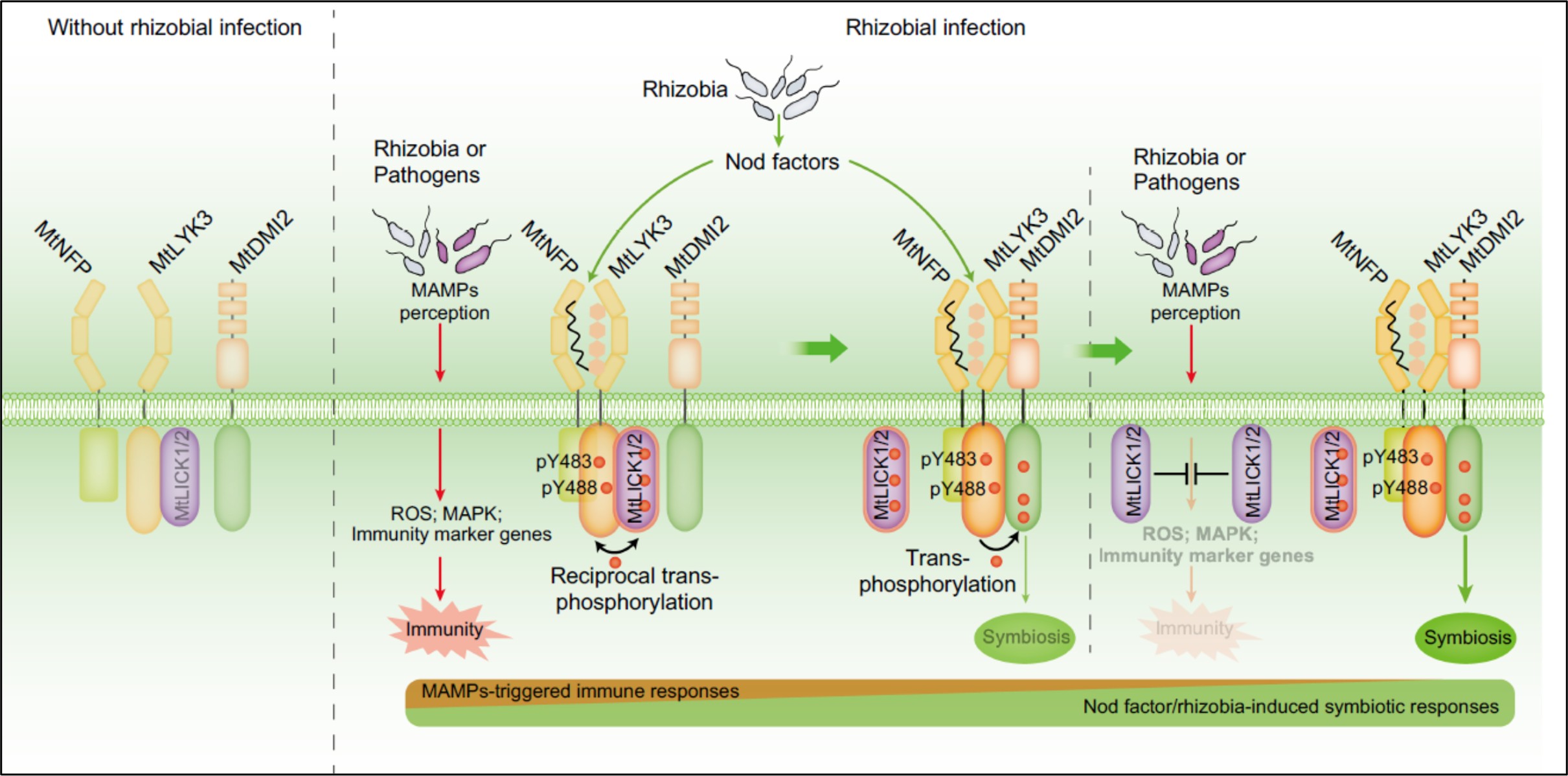
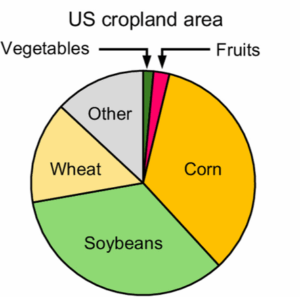
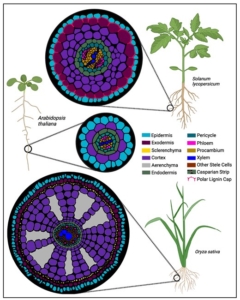 The May 2025 issue of The Plant Cell has a focus on Arabidopsis and how this simple model plant has had an outsized impact on our understanding of pretty much everything botanical (and beyond). Alongside several articles highlighting this impact, this review by Roeder et al. features eight short vignettes on topics that do not translate from Arabidopsis. If you are at all familiar with Arabidopsis, you can probably guess what a few of these are; the first vignette highlights the incredibly easy and efficient floral dip method for transforming Arabidopsis, and discusses why it doesn’t work for most plants. Other vignettes cover Arabidopsis’ lack of mycorrhizal associations and C4 photosynthesis; challenges of orthology and transcriptional regulatory region mapping to bigger genomes; the limitations of very simple root anatomy of Arabidopsis; challenges of studying drought responses in Arabidopsis; and finally, differences in circadian biology between Arabidopsis and other plants. It’s a fascinating and illuminating article, and should be useful to those who seek to understand the strengths and weaknesses of working with model systems. (Summary by Mary Williams @PlantTeaching.bsky.social) Plant Cell 10.1093/plcell/koaf036
The May 2025 issue of The Plant Cell has a focus on Arabidopsis and how this simple model plant has had an outsized impact on our understanding of pretty much everything botanical (and beyond). Alongside several articles highlighting this impact, this review by Roeder et al. features eight short vignettes on topics that do not translate from Arabidopsis. If you are at all familiar with Arabidopsis, you can probably guess what a few of these are; the first vignette highlights the incredibly easy and efficient floral dip method for transforming Arabidopsis, and discusses why it doesn’t work for most plants. Other vignettes cover Arabidopsis’ lack of mycorrhizal associations and C4 photosynthesis; challenges of orthology and transcriptional regulatory region mapping to bigger genomes; the limitations of very simple root anatomy of Arabidopsis; challenges of studying drought responses in Arabidopsis; and finally, differences in circadian biology between Arabidopsis and other plants. It’s a fascinating and illuminating article, and should be useful to those who seek to understand the strengths and weaknesses of working with model systems. (Summary by Mary Williams @PlantTeaching.bsky.social) Plant Cell 10.1093/plcell/koaf036