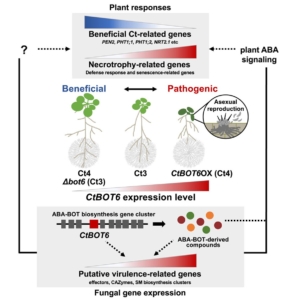
Forum. Plant surveillance: The emerging role of substrate-binding proteins
 Plants are sessile organisms and, unlike animals, cannot escape adverse environmental conditions. To cope with this limitation, they have evolved a complex surveillance system to detect and respond to fluctuating conditions such as resource scarcity, environmental changes, and pathogen attacks. Membrane-bound receptors play a central role in these processes. It has long been recognized that plants possess a significantly larger repertoire of receptor-like kinases (RLKs) and receptor-like proteins (RLPs) compared to their mammalian counterparts—an adaptation thought to compensate for their lack of mobility. Traditionally, signal transduction in plants has been viewed through a relatively simple lens, primarily involving direct ligand–receptor interactions. In this new work, Pei and colleagues discuss new possibilities inspired by the mammalian system, and an emerging multi-functional RLK in plants – FERONIA. In many cases, substrate-binding proteins (SBPs) may act as intermediaries between ligands and receptors, offering structural and spatial flexibility as well as the ability to integrate multiple signals. This forum article presents new perspectives on unraveling the molecular mechanisms of signal transduction in plants, highlighting the critical roles of RLKs, RLPs, and their associated components. (Summary by Ching Chan @ntnuchanlab) Trends Plant Sci. 10.1016/j.tplants.2025.03.018
Plants are sessile organisms and, unlike animals, cannot escape adverse environmental conditions. To cope with this limitation, they have evolved a complex surveillance system to detect and respond to fluctuating conditions such as resource scarcity, environmental changes, and pathogen attacks. Membrane-bound receptors play a central role in these processes. It has long been recognized that plants possess a significantly larger repertoire of receptor-like kinases (RLKs) and receptor-like proteins (RLPs) compared to their mammalian counterparts—an adaptation thought to compensate for their lack of mobility. Traditionally, signal transduction in plants has been viewed through a relatively simple lens, primarily involving direct ligand–receptor interactions. In this new work, Pei and colleagues discuss new possibilities inspired by the mammalian system, and an emerging multi-functional RLK in plants – FERONIA. In many cases, substrate-binding proteins (SBPs) may act as intermediaries between ligands and receptors, offering structural and spatial flexibility as well as the ability to integrate multiple signals. This forum article presents new perspectives on unraveling the molecular mechanisms of signal transduction in plants, highlighting the critical roles of RLKs, RLPs, and their associated components. (Summary by Ching Chan @ntnuchanlab) Trends Plant Sci. 10.1016/j.tplants.2025.03.018
Perspective. The art of drying gracefully: The future for desiccation research
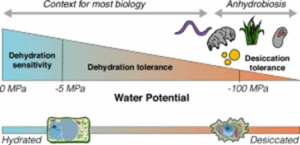 In their perspective article, “Life on the dry side: a roadmap to understanding desiccation tolerance and accelerating translational applications,” Marks et al. outline the current state and future promise of desiccation tolerance research. They define desiccation tolerance as “the ability to dry to a quiescent state and resume normal cellular function when rehydrated,” a process fundamentally distinct from drought resistance or avoidance. Tracing its evolutionary origins to ancestral prokaryotic, archaeal, and algal lineages, desiccation tolerance has emerged multiple times across diverse taxa, often accompanied by convergent molecular adaptations, such as the repeated evolution of Early Light Inducible Proteins (ELIPs) in vascular plants. The ecological breadth of desiccation tolerant organisms is similarly diverse, with species inhabiting both arid and intermittently wet environments, frequently exhibiting tolerance to other extreme conditions. Physiologically, desiccation tolerance is characterized by intricate cellular responses, including vitrification of the cytosol, reactive oxygen species (ROS) accumulation, and structural modifications like leaf curling. However, progress in the field is hindered by inconsistent experimental frameworks. The authors advocate for standardized protocols that include precise hydration status measurements, viability assays post-rehydration, and time-course studies using indicators such as chlorophyll fluorescence. Microscopy, if properly executed to prevent sample rehydration, also offers key insights. Emphasizing the importance of FAIR (Findable, Accessible, Interoperable, and Reusable) data principles, the authors argue that collaborative and unified research practices will be crucial to unlocking DT’s potential in medicine, agriculture, and industry, especially for supporting smallholder farmers and advancing biotechnological innovation. (Summary by Stephanie Temnyk @STemnyk) Nature Communications 10.1038/s41467-025-58656-y
In their perspective article, “Life on the dry side: a roadmap to understanding desiccation tolerance and accelerating translational applications,” Marks et al. outline the current state and future promise of desiccation tolerance research. They define desiccation tolerance as “the ability to dry to a quiescent state and resume normal cellular function when rehydrated,” a process fundamentally distinct from drought resistance or avoidance. Tracing its evolutionary origins to ancestral prokaryotic, archaeal, and algal lineages, desiccation tolerance has emerged multiple times across diverse taxa, often accompanied by convergent molecular adaptations, such as the repeated evolution of Early Light Inducible Proteins (ELIPs) in vascular plants. The ecological breadth of desiccation tolerant organisms is similarly diverse, with species inhabiting both arid and intermittently wet environments, frequently exhibiting tolerance to other extreme conditions. Physiologically, desiccation tolerance is characterized by intricate cellular responses, including vitrification of the cytosol, reactive oxygen species (ROS) accumulation, and structural modifications like leaf curling. However, progress in the field is hindered by inconsistent experimental frameworks. The authors advocate for standardized protocols that include precise hydration status measurements, viability assays post-rehydration, and time-course studies using indicators such as chlorophyll fluorescence. Microscopy, if properly executed to prevent sample rehydration, also offers key insights. Emphasizing the importance of FAIR (Findable, Accessible, Interoperable, and Reusable) data principles, the authors argue that collaborative and unified research practices will be crucial to unlocking DT’s potential in medicine, agriculture, and industry, especially for supporting smallholder farmers and advancing biotechnological innovation. (Summary by Stephanie Temnyk @STemnyk) Nature Communications 10.1038/s41467-025-58656-y
Review: Translating research from Arabidopsis to crops
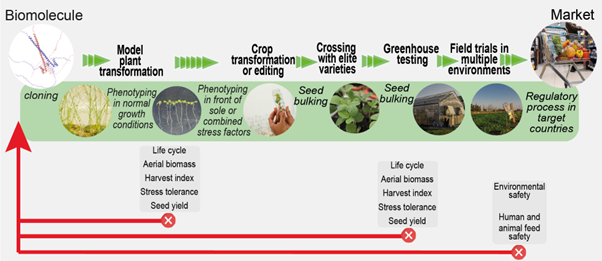 This year marks the 25th anniversary of the publication of the Arabidopsis genome, a milestone that lives large in those of us who experienced it and eagerly envisioned how this new knowledge would be used. The May 2025 issue of The Plant Cell focuses on how research on this little model plant has translated outwards. One of the reviews in this focus issue, by Uauy et al., looks closely at the challenges and opportunities for crop improvement that stem from Arabidopsis research. This review follows several genes, correlated with a given trait in Arabidopsis, on their journey into crop plants. The highlighted genes include those associated with stress tolerance, production of omega-3 fatty acids, starch synthesis, and epidermal patterning / cuticle formation. Each story is interesting on its own, but when read together several key messages emerge. Translating from a simple model species is difficult but vital to crop research, due to the ease with which gene expression and function can be manipulated and analyzed in Arabidopsis. However, obstacles to translation include challenges of crop plant transformation, differences in the fundamental biology of Arabidopsis plants versus crop plants, and the fact that field-grown crops experience tremendous environmental fluctuations that aren’t usually accounted for in Arabidopsis studies. Enjoy this review and the rest of the focus issue, and join us May 30th for a lively panel discussion with some of the authors whose work appears in it (register here). (Summary by Mary Williams @PlantTeaching.bsky.social) Plant Cell 10.1093/plcell/koaf059
This year marks the 25th anniversary of the publication of the Arabidopsis genome, a milestone that lives large in those of us who experienced it and eagerly envisioned how this new knowledge would be used. The May 2025 issue of The Plant Cell focuses on how research on this little model plant has translated outwards. One of the reviews in this focus issue, by Uauy et al., looks closely at the challenges and opportunities for crop improvement that stem from Arabidopsis research. This review follows several genes, correlated with a given trait in Arabidopsis, on their journey into crop plants. The highlighted genes include those associated with stress tolerance, production of omega-3 fatty acids, starch synthesis, and epidermal patterning / cuticle formation. Each story is interesting on its own, but when read together several key messages emerge. Translating from a simple model species is difficult but vital to crop research, due to the ease with which gene expression and function can be manipulated and analyzed in Arabidopsis. However, obstacles to translation include challenges of crop plant transformation, differences in the fundamental biology of Arabidopsis plants versus crop plants, and the fact that field-grown crops experience tremendous environmental fluctuations that aren’t usually accounted for in Arabidopsis studies. Enjoy this review and the rest of the focus issue, and join us May 30th for a lively panel discussion with some of the authors whose work appears in it (register here). (Summary by Mary Williams @PlantTeaching.bsky.social) Plant Cell 10.1093/plcell/koaf059
Rewiring sugar signaling for global agriculture: A decade of progress in harnessing the T6P pathway
 While sugar signaling plays a crucial role in grain filling and ultimately determines grain crop yield, direct genetic manipulation often results in pleiotropic effects and raises legislative concerns, limiting its practical application in agriculture. To address this, a “signaling-precursor” concept was introduced about a decade ago, involving the use of membrane-permeable analogues of trehalose-6-phosphate (T6P) to enhance plant performance. Unlike genetic modification, the external application of T6P analogues circumvents regulatory barriers and allows for flexible timing. In laboratory settings, such treatments have been shown to increase grain number, sugar content, and resilience to dehydration in both Arabidopsis and wheat. In a recent study, Griffiths and colleagues of the same research group validated the potential of this approach under field conditions. Three wheat cultivars were grown in field trials over four years, and their performance under dehydration stress was assessed. Remarkably, chemical treatment enhanced both source processes (CO₂ fixation and energy production) and sink processes (starch biosynthesis and endosperm enlargement), resulting in a 9–22% increase in wheat yield. Moreover, this technology is also effective in other cereals, such as sorghum and barley, underscoring the broad application potential of targeting the T6P signaling pathway in agriculture. (Summary by Ching Chan @ntnuchanlab) Nature Biotechnology 10.1038/s41587-025-02611-1
While sugar signaling plays a crucial role in grain filling and ultimately determines grain crop yield, direct genetic manipulation often results in pleiotropic effects and raises legislative concerns, limiting its practical application in agriculture. To address this, a “signaling-precursor” concept was introduced about a decade ago, involving the use of membrane-permeable analogues of trehalose-6-phosphate (T6P) to enhance plant performance. Unlike genetic modification, the external application of T6P analogues circumvents regulatory barriers and allows for flexible timing. In laboratory settings, such treatments have been shown to increase grain number, sugar content, and resilience to dehydration in both Arabidopsis and wheat. In a recent study, Griffiths and colleagues of the same research group validated the potential of this approach under field conditions. Three wheat cultivars were grown in field trials over four years, and their performance under dehydration stress was assessed. Remarkably, chemical treatment enhanced both source processes (CO₂ fixation and energy production) and sink processes (starch biosynthesis and endosperm enlargement), resulting in a 9–22% increase in wheat yield. Moreover, this technology is also effective in other cereals, such as sorghum and barley, underscoring the broad application potential of targeting the T6P signaling pathway in agriculture. (Summary by Ching Chan @ntnuchanlab) Nature Biotechnology 10.1038/s41587-025-02611-1
Boosting C4 photosynthesis and productivity by elevating Rubisco levels in sorghum and sugarcane
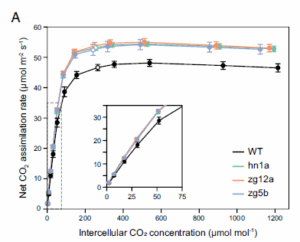 The United Nations projects that by 2050, global food production must increase by 60% to meet growing demand, a goal that must be met under the pressures of global climate change and without further agricultural land expansion. With rising atmospheric CO₂ levels, Rubisco has emerged as the primary limitation to light-saturated leaf CO₂ assimilation rates (Aₛₐₜ) in C₄ crops, which represent a significant share of global agricultural output. Salesse-Smith et al. overexpressed the Rubisco small subunit (RbcS) alongside Rubisco accumulation factor 1 (Raf1) genes in both sorghum and sugarcane, increasing their photosynthetic efficiency and productivity. The authors reported that transgenic upregulation of RbcS and Raf1 led to a 13-25% increase in Rubisco abundance in sorghum and up to 90% in sugarcane. Additionally, co-upregulation of RbcS and Raf1 correlated with increased maximum in vivo Rubisco carboxylation rates (Vc,max), CO2 assimilation, and photosynthetic light induction. For both species, Aₛₐₜ increased by 12–15%, and Rubisco enzyme activity rose by approximately 40%. In sorghum, these changes also led to faster photosynthetic induction and reduced bundle sheath leakiness. A field trial in sorghum confirmed these results, which translated into a 15.5% increase in biomass accumulation, while greenhouse-grown sugarcane showed a 37% to 81% increase. This study presents valuable means to enhance the productivity of these C4 crops as global atmospheric conditions evolve and sets the ground for testing Rubisco upregulation in elite cultivars and hybrids in multilocation and multiyear yield trials. (Summary by Elisa De Meo, www.linkedin.com/in/elisa-de-meo-25415a20b) PNAS, 10.1073/pnas.2419943122
The United Nations projects that by 2050, global food production must increase by 60% to meet growing demand, a goal that must be met under the pressures of global climate change and without further agricultural land expansion. With rising atmospheric CO₂ levels, Rubisco has emerged as the primary limitation to light-saturated leaf CO₂ assimilation rates (Aₛₐₜ) in C₄ crops, which represent a significant share of global agricultural output. Salesse-Smith et al. overexpressed the Rubisco small subunit (RbcS) alongside Rubisco accumulation factor 1 (Raf1) genes in both sorghum and sugarcane, increasing their photosynthetic efficiency and productivity. The authors reported that transgenic upregulation of RbcS and Raf1 led to a 13-25% increase in Rubisco abundance in sorghum and up to 90% in sugarcane. Additionally, co-upregulation of RbcS and Raf1 correlated with increased maximum in vivo Rubisco carboxylation rates (Vc,max), CO2 assimilation, and photosynthetic light induction. For both species, Aₛₐₜ increased by 12–15%, and Rubisco enzyme activity rose by approximately 40%. In sorghum, these changes also led to faster photosynthetic induction and reduced bundle sheath leakiness. A field trial in sorghum confirmed these results, which translated into a 15.5% increase in biomass accumulation, while greenhouse-grown sugarcane showed a 37% to 81% increase. This study presents valuable means to enhance the productivity of these C4 crops as global atmospheric conditions evolve and sets the ground for testing Rubisco upregulation in elite cultivars and hybrids in multilocation and multiyear yield trials. (Summary by Elisa De Meo, www.linkedin.com/in/elisa-de-meo-25415a20b) PNAS, 10.1073/pnas.2419943122
Leaf senescence mapped through Arabidopsis single-nucleus RNA-Seq atlas
 As both the power and accessibility of single-cell RNA sequencing increases, more reference datasets have become available, documenting the transcriptome within a variety of model plants. While invaluable resources that have pushed science forward, these datasets are often limited to single tissues or few timepoints. Additionally, they lack information on tissues like senescing leaves, flowers, and fruits because it is more difficult to isolate single-cell protoplasts from these tissues. To have a fuller understanding of the interconnected spatiotemporal relationships throughout development, Guo et al. have constructed an atlas of Arabidopsis single-nucleus RNA sequencing transcriptomic information across 20 tissues at multiple developmental stages from vegetative growth to reproductive growth, identifying 38 cell types. This atlas focuses on characterizing leaf senescence and nutrient allocation, two topics neglected by previous methods. Key regulators of senescence were identified by the atlas and developed into a quantitative leaf aging index that can assess age state of leaf cells from single-cell transcriptomic information. 6,591 senescence-associated genes and 1,875 youth-associated genes were identified, and aging hub genes were determined by co-expression network analysis. The atlas was also applied to nutrient allocation, mapping the carbon and nitrogen transporters involved in source-sink dynamics. In the future, this dataset could be used to characterize gene interactions between the key regulator genes and associated signaling pathways, integrating the dynamics of senescence and nutrient source-sink allocation. (Summary by Elise Krespan) Cell https://doi.org/10.1016/j.cell.2025.03.024
As both the power and accessibility of single-cell RNA sequencing increases, more reference datasets have become available, documenting the transcriptome within a variety of model plants. While invaluable resources that have pushed science forward, these datasets are often limited to single tissues or few timepoints. Additionally, they lack information on tissues like senescing leaves, flowers, and fruits because it is more difficult to isolate single-cell protoplasts from these tissues. To have a fuller understanding of the interconnected spatiotemporal relationships throughout development, Guo et al. have constructed an atlas of Arabidopsis single-nucleus RNA sequencing transcriptomic information across 20 tissues at multiple developmental stages from vegetative growth to reproductive growth, identifying 38 cell types. This atlas focuses on characterizing leaf senescence and nutrient allocation, two topics neglected by previous methods. Key regulators of senescence were identified by the atlas and developed into a quantitative leaf aging index that can assess age state of leaf cells from single-cell transcriptomic information. 6,591 senescence-associated genes and 1,875 youth-associated genes were identified, and aging hub genes were determined by co-expression network analysis. The atlas was also applied to nutrient allocation, mapping the carbon and nitrogen transporters involved in source-sink dynamics. In the future, this dataset could be used to characterize gene interactions between the key regulator genes and associated signaling pathways, integrating the dynamics of senescence and nutrient source-sink allocation. (Summary by Elise Krespan) Cell https://doi.org/10.1016/j.cell.2025.03.024
Identification of the final set of Mendel’s pea genes
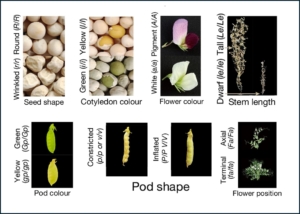 We can be assured that every biology student is exposed to plants at least once, when they learn about the genetic laws that Mendel formulated from his studies on peas. His classic work focused on seven traits that exhibited simple dominant-recessive characteristics, such as seed color and shape and flower color. Of these seven traits, the genes controlling four were previously identified. This exciting new work by Feng et al. reveals the identity of the final three genes, controlling pod color, pod form, and fasciation (fasciation describes a growth condition where the stem or inflorescence forms as a flattened band). The authors explored a diversity panel of 697 isolates from four species of Pisum, creating a genomic variation map, and they scored each for the seven traits that Mendel worked with, thus confirming the identity of the four previously known genes, and identifying candidates for the three remaining unknown genes. It’s a fascinating article and one that would be exciting to share with students in an advanced genetics class. Or, with less advanced students, skip right to the Research Briefing for the author’s more accessible summary. (Summary by Mary Williams @PlantTeaching.bsky.social) Nature 10.1038/s41586-025-08891-6, Research Briefing https://www.nature.com/articles/d41586-025-01164-2
We can be assured that every biology student is exposed to plants at least once, when they learn about the genetic laws that Mendel formulated from his studies on peas. His classic work focused on seven traits that exhibited simple dominant-recessive characteristics, such as seed color and shape and flower color. Of these seven traits, the genes controlling four were previously identified. This exciting new work by Feng et al. reveals the identity of the final three genes, controlling pod color, pod form, and fasciation (fasciation describes a growth condition where the stem or inflorescence forms as a flattened band). The authors explored a diversity panel of 697 isolates from four species of Pisum, creating a genomic variation map, and they scored each for the seven traits that Mendel worked with, thus confirming the identity of the four previously known genes, and identifying candidates for the three remaining unknown genes. It’s a fascinating article and one that would be exciting to share with students in an advanced genetics class. Or, with less advanced students, skip right to the Research Briefing for the author’s more accessible summary. (Summary by Mary Williams @PlantTeaching.bsky.social) Nature 10.1038/s41586-025-08891-6, Research Briefing https://www.nature.com/articles/d41586-025-01164-2
Unraveling the epitranscriptomic-chromatin axis: How NERD regulates flowering in Arabidopsis
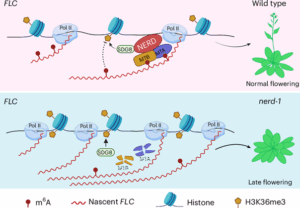 Flowering in Arabidopsis thaliana is governed by complex genetic and epigenetic networks. A central component of this regulation is the floral repressor FLOWERING LOCUS C (FLC), whose expression is modulated by histone modifications, particularly the activating H3K36me3 and the repressive H3K27me3 marks. While these chromatin-level mechanisms have been extensively studied, emerging evidence highlights a crucial role of RNA modifications, collectively termed epitranscriptomics, in fine-tuning gene expression. A recent study by Shao et al. uncovers a pivotal function for the plant-specific CCCH-type zinc finger protein NERD (NEEDED FOR RDR2-INDEPENDENT DNA METHYLATION) in modulating N6-methyladenosine (m⁶A) deposition on FLC transcripts. The authors show that NERD physically interacts with core components of the m⁶A methyltransferase complex (MTA and MTB) to promote m⁶A modification of nascent RNA. This N6-methyladenosine (m⁶A) deposition is known to act as a transcriptional repressor. Through mass spectrometry and m⁶A RNA immunoprecipitation sequencing, the study reveals that nerd-1 mutants exhibit globally reduced m⁶A levels in both nuclear and mature mRNAs. Intriguingly, NERD interacts with the H3K36me3 methyltransferase SET DOMAIN GROUP 8 (SDG8), establishing a mechanistic link between RNA methylation and chromatin regulation. Loss of NERD function leads to increased levels of H3K36me3 at FLC, correlating with elevated transcription of this floral repressor and delayed flowering. This study uncovers a novel epitranscriptomic–chromatin regulatory axis in flowering control, highlighting the intricate cross-talk between RNA and chromatin landscapes and opening new avenues for exploring RNA-based gene regulations in plant development. (Summary by Gourav Arora @gouravarora.bsky.social) Nature Plants 10.1038/s41477-025-01945-7
Flowering in Arabidopsis thaliana is governed by complex genetic and epigenetic networks. A central component of this regulation is the floral repressor FLOWERING LOCUS C (FLC), whose expression is modulated by histone modifications, particularly the activating H3K36me3 and the repressive H3K27me3 marks. While these chromatin-level mechanisms have been extensively studied, emerging evidence highlights a crucial role of RNA modifications, collectively termed epitranscriptomics, in fine-tuning gene expression. A recent study by Shao et al. uncovers a pivotal function for the plant-specific CCCH-type zinc finger protein NERD (NEEDED FOR RDR2-INDEPENDENT DNA METHYLATION) in modulating N6-methyladenosine (m⁶A) deposition on FLC transcripts. The authors show that NERD physically interacts with core components of the m⁶A methyltransferase complex (MTA and MTB) to promote m⁶A modification of nascent RNA. This N6-methyladenosine (m⁶A) deposition is known to act as a transcriptional repressor. Through mass spectrometry and m⁶A RNA immunoprecipitation sequencing, the study reveals that nerd-1 mutants exhibit globally reduced m⁶A levels in both nuclear and mature mRNAs. Intriguingly, NERD interacts with the H3K36me3 methyltransferase SET DOMAIN GROUP 8 (SDG8), establishing a mechanistic link between RNA methylation and chromatin regulation. Loss of NERD function leads to increased levels of H3K36me3 at FLC, correlating with elevated transcription of this floral repressor and delayed flowering. This study uncovers a novel epitranscriptomic–chromatin regulatory axis in flowering control, highlighting the intricate cross-talk between RNA and chromatin landscapes and opening new avenues for exploring RNA-based gene regulations in plant development. (Summary by Gourav Arora @gouravarora.bsky.social) Nature Plants 10.1038/s41477-025-01945-7
Cracking the code of rice grain quality under heat stress
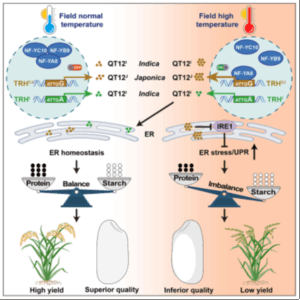 Rice is one of the most important food crops globally, providing more than 20% of the world’s calorie intake and over 75% for the population in Asia. Therefore, apart from yield, grain quality is another critical agronomic trait for breeding and improvement, especially under stressful environmental conditions. By comparing the two major rice subspecies, Indica and Japonica, which exhibit differential responses to heat stress (Indica is more thermotolerant), Li and colleagues identified a molecular “switch” – QT12 – that regulates grain chalkiness, a well-established indicator of grain development and quality. Through comprehensive molecular and biochemical analyses, the researchers found that grain quailty thermosensitivity occurs when QT12 expression increases at higher temperatures. In varities with thermotolerant grain quality, QT12 expression does not increase with increasing heat, and this heat-sensitive expression is associated with Nuclear Factor Y (NF-Y) proteins. QT12 modulates the balance of storage protein and starch content during grain filling, ultimately affecting overall grain quality. Interestingly, although located on different chromosomes, QT12 and NF-Ys have been co-selected during evolution. Distinct trait regulatory haplotypes (TRHs) are identified in heat-tolerant and heat-sensitive varieties, offering valuable tools for marker-assisted breeding. (Summary by Ching Chan @ntnuchanlab) Cell 10.1016/j.cell.2025.04.011
Rice is one of the most important food crops globally, providing more than 20% of the world’s calorie intake and over 75% for the population in Asia. Therefore, apart from yield, grain quality is another critical agronomic trait for breeding and improvement, especially under stressful environmental conditions. By comparing the two major rice subspecies, Indica and Japonica, which exhibit differential responses to heat stress (Indica is more thermotolerant), Li and colleagues identified a molecular “switch” – QT12 – that regulates grain chalkiness, a well-established indicator of grain development and quality. Through comprehensive molecular and biochemical analyses, the researchers found that grain quailty thermosensitivity occurs when QT12 expression increases at higher temperatures. In varities with thermotolerant grain quality, QT12 expression does not increase with increasing heat, and this heat-sensitive expression is associated with Nuclear Factor Y (NF-Y) proteins. QT12 modulates the balance of storage protein and starch content during grain filling, ultimately affecting overall grain quality. Interestingly, although located on different chromosomes, QT12 and NF-Ys have been co-selected during evolution. Distinct trait regulatory haplotypes (TRHs) are identified in heat-tolerant and heat-sensitive varieties, offering valuable tools for marker-assisted breeding. (Summary by Ching Chan @ntnuchanlab) Cell 10.1016/j.cell.2025.04.011
Training scientists to make their data FAIR
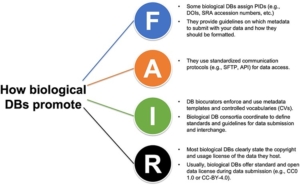 I’m sure many of you have experienced frustration when trying to access an intriguing dataset that either doesn’t exist, isn’t open, or is set up in an impossibly unintuitive manner. Part of that problem stems from a lack of training of early-career scientists in how to make their data findable, accessible, interoperable, and reusable (FAIR). Thankfully, Maranno and other members of the AgBioData Education working group have created a free, online curriculum to support the training of undergraduate and graduate students. The curriculum includes seven lesson modules and was designed around core concepts and specific learning objectives needed to use databases following the FAIR principles. These lessons and their objectives are described in this manuscript, which gives educators an overview of the key ideas covered in the curriculum. The lessons themselves are housed and freely available on the AgBioData Zenodo community https://zenodo.org/records/14278084. They have also developed a Curriculum Implementation/Development Program (https://www.agbiodata.org/agb-education-implement-awards) to financially support individuals who wish to implement and/or contribute to the curriculum; check it out. This is an excellent, much needed, freely accessible resource that will help all who seek, use, teach, or want to learn about databases. (Summary by Mary Williams @PlantTeaching.bsky.social) Database 10.1093/database/baaf034
I’m sure many of you have experienced frustration when trying to access an intriguing dataset that either doesn’t exist, isn’t open, or is set up in an impossibly unintuitive manner. Part of that problem stems from a lack of training of early-career scientists in how to make their data findable, accessible, interoperable, and reusable (FAIR). Thankfully, Maranno and other members of the AgBioData Education working group have created a free, online curriculum to support the training of undergraduate and graduate students. The curriculum includes seven lesson modules and was designed around core concepts and specific learning objectives needed to use databases following the FAIR principles. These lessons and their objectives are described in this manuscript, which gives educators an overview of the key ideas covered in the curriculum. The lessons themselves are housed and freely available on the AgBioData Zenodo community https://zenodo.org/records/14278084. They have also developed a Curriculum Implementation/Development Program (https://www.agbiodata.org/agb-education-implement-awards) to financially support individuals who wish to implement and/or contribute to the curriculum; check it out. This is an excellent, much needed, freely accessible resource that will help all who seek, use, teach, or want to learn about databases. (Summary by Mary Williams @PlantTeaching.bsky.social) Database 10.1093/database/baaf034
 Rice is one of the most important food crops globally, providing more than 20% of the world’s calorie intake and over 75% for the population in Asia. Therefore, apart from yield, grain quality is another critical agronomic trait for breeding and improvement, especially under stressful environmental conditions. By comparing the two major rice subspecies, Indica and Japonica, which exhibit differential responses to heat stress (Indica is more thermotolerant), Li and colleagues identified a molecular “switch” – QT12 – that regulates grain chalkiness, a well-established indicator of grain development and quality. Through comprehensive molecular and biochemical analyses, the researchers found that grain quailty thermosensitivity occurs when QT12 expression increases at higher temperatures. In varities with thermotolerant grain quality, QT12 expression does not increase with increasing heat, and this heat-sensitive expression is associated with Nuclear Factor Y (NF-Y) proteins. QT12 modulates the balance of storage protein and starch content during grain filling, ultimately affecting overall grain quality. Interestingly, although located on different chromosomes, QT12 and NF-Ys have been co-selected during evolution. Distinct trait regulatory haplotypes (TRHs) are identified in heat-tolerant and heat-sensitive varieties, offering valuable tools for marker-assisted breeding. (Summary by Ching Chan @ntnuchanlab) Cell 10.1016/j.cell.2025.04.011
Rice is one of the most important food crops globally, providing more than 20% of the world’s calorie intake and over 75% for the population in Asia. Therefore, apart from yield, grain quality is another critical agronomic trait for breeding and improvement, especially under stressful environmental conditions. By comparing the two major rice subspecies, Indica and Japonica, which exhibit differential responses to heat stress (Indica is more thermotolerant), Li and colleagues identified a molecular “switch” – QT12 – that regulates grain chalkiness, a well-established indicator of grain development and quality. Through comprehensive molecular and biochemical analyses, the researchers found that grain quailty thermosensitivity occurs when QT12 expression increases at higher temperatures. In varities with thermotolerant grain quality, QT12 expression does not increase with increasing heat, and this heat-sensitive expression is associated with Nuclear Factor Y (NF-Y) proteins. QT12 modulates the balance of storage protein and starch content during grain filling, ultimately affecting overall grain quality. Interestingly, although located on different chromosomes, QT12 and NF-Ys have been co-selected during evolution. Distinct trait regulatory haplotypes (TRHs) are identified in heat-tolerant and heat-sensitive varieties, offering valuable tools for marker-assisted breeding. (Summary by Ching Chan @ntnuchanlab) Cell 10.1016/j.cell.2025.04.011