Review: Cycad, chemicals, and coevolution
 Cycads are an ancient lineage of gymnosperms with fascinating ecological interactions. In a recent review, Salzman et al. examine the various adaptations of cycads, from attracting pollinators to repelling parasites, focusing on the roles of their wide array of specialized metabolites. A somewhat unique trait in gymnosperms, most cycads have obligate insect pollination. To attract pollinators, cycads release a wide array of volatile organic compounds (VOCs) in their cones. As dioecious plants, they utilize daily fluctuations of VOCs to attract pollinators to their male cones to gather pollen, then repel them to encourage them towards female cones. Certain genera of cycads are thermogenic, with the ability to generate heat in their cones to attract insect pollinators. Aside from pollinator interactions, cycads also produce a myriad of phytotoxins in their foliage, which has driven tight coevolutionary relationships with specialized insects that have adapted to withstand these chemical defenses. Their use of secondary metabolites extends to below ground tissue where they secrete metabolites to attract nitrogen-fixing mutualists and boost their metabolic activity. Because they retain many ancient traits not found in most gymnosperms, cycads present a valuable opportunity to help bridge the gap between living and early seed plants, offering key insights into the evolutionary history of plant-animal interactions. (Summary by Xavier Ozowara [email protected]). New Phytologist https://doi.org/10.1111/nph.70109
Cycads are an ancient lineage of gymnosperms with fascinating ecological interactions. In a recent review, Salzman et al. examine the various adaptations of cycads, from attracting pollinators to repelling parasites, focusing on the roles of their wide array of specialized metabolites. A somewhat unique trait in gymnosperms, most cycads have obligate insect pollination. To attract pollinators, cycads release a wide array of volatile organic compounds (VOCs) in their cones. As dioecious plants, they utilize daily fluctuations of VOCs to attract pollinators to their male cones to gather pollen, then repel them to encourage them towards female cones. Certain genera of cycads are thermogenic, with the ability to generate heat in their cones to attract insect pollinators. Aside from pollinator interactions, cycads also produce a myriad of phytotoxins in their foliage, which has driven tight coevolutionary relationships with specialized insects that have adapted to withstand these chemical defenses. Their use of secondary metabolites extends to below ground tissue where they secrete metabolites to attract nitrogen-fixing mutualists and boost their metabolic activity. Because they retain many ancient traits not found in most gymnosperms, cycads present a valuable opportunity to help bridge the gap between living and early seed plants, offering key insights into the evolutionary history of plant-animal interactions. (Summary by Xavier Ozowara [email protected]). New Phytologist https://doi.org/10.1111/nph.70109
A plant kinesin-microtubule module governs chromosome alignment in mitosis
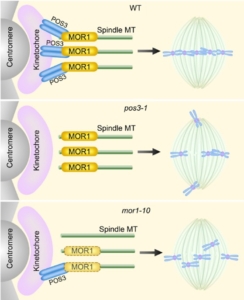 During cell division, it’s essential that each new cell receives a complete set of chromosomes. To ensure this, chromosomes first align at the center of the cell, where structures called microtubules connect to a region on each chromosome known as the centromere. This alignment step—called chromosome congression—is crucial for proper chromosome separation. In this study, Chen et al. used a chemical genetic screen in Arabidopsis thaliana to identify mutants that are hypersensitive to a drug (propyzamide) that disrupts microtubules. One of these mutants, named propyzamide oversensitive3-1 (pos3-1), was found to have defects in cell division. The mutated gene encodes POS3, a motor protein that shares features with an animal protein called CENP-E, which helps chromosomes align during mitosis. The authors show that POS3 binds to microtubules and dynamically localizes to the kinetochores—the protein structures on chromosomes where microtubules attach. Loss of POS3 results in delayed mitosis, improper alignment of chromosomes, and errors in chromosome number in daughter cells (a condition called aneuploidy). Unexpectedly, the researchers discovered that POS3 physically interacts with MOR1, a microtubule polymerase previously known to support microtubule growth and organization. MOR1 helps POS3 localize correctly to the kinetochore. When both POS3 and MOR1 are mutated, cells show even more severe division errors, indicating these two proteins work together to coordinate chromosome alignment and segregation. This work uncovers a plant-specific system involving a kinesin motor (POS3) and a microtubule-regulating protein (MOR1) that together ensure accurate chromosome behavior during mitosis. It advances our understanding of how plants organize their mitotic machinery in the absence of structures like centrosomes, which are present in animal cells. (Summary by Muhammad Aamir Khan @MAKNature1998) Plant Cell 10.1093/plcell/koaf053
During cell division, it’s essential that each new cell receives a complete set of chromosomes. To ensure this, chromosomes first align at the center of the cell, where structures called microtubules connect to a region on each chromosome known as the centromere. This alignment step—called chromosome congression—is crucial for proper chromosome separation. In this study, Chen et al. used a chemical genetic screen in Arabidopsis thaliana to identify mutants that are hypersensitive to a drug (propyzamide) that disrupts microtubules. One of these mutants, named propyzamide oversensitive3-1 (pos3-1), was found to have defects in cell division. The mutated gene encodes POS3, a motor protein that shares features with an animal protein called CENP-E, which helps chromosomes align during mitosis. The authors show that POS3 binds to microtubules and dynamically localizes to the kinetochores—the protein structures on chromosomes where microtubules attach. Loss of POS3 results in delayed mitosis, improper alignment of chromosomes, and errors in chromosome number in daughter cells (a condition called aneuploidy). Unexpectedly, the researchers discovered that POS3 physically interacts with MOR1, a microtubule polymerase previously known to support microtubule growth and organization. MOR1 helps POS3 localize correctly to the kinetochore. When both POS3 and MOR1 are mutated, cells show even more severe division errors, indicating these two proteins work together to coordinate chromosome alignment and segregation. This work uncovers a plant-specific system involving a kinesin motor (POS3) and a microtubule-regulating protein (MOR1) that together ensure accurate chromosome behavior during mitosis. It advances our understanding of how plants organize their mitotic machinery in the absence of structures like centrosomes, which are present in animal cells. (Summary by Muhammad Aamir Khan @MAKNature1998) Plant Cell 10.1093/plcell/koaf053
Breaking the breeding cycle: Parthenogenesis paves the way for faster sunflower breeding
 Hybrid crops are key to modern agriculture, offering higher yields, improved stress resilience, and greater uniformity through heterosis (“hybrid vigor”), where offspring from two different inbred lines outperform their parents. However, producing homozygous inbred lines for stable hybrid seed production traditionally takes at least six generations of self-pollination, slowing breeding progress. Doubled haploid (DH) technology offers a faster solution by generating homozygous lines in a single generation through haploid embryo formation and genome doubling. While DH systems are established in maize, their use in other crops has been limited by inefficiency and genotype dependence. In a serendipitous discovery, Lv et al. found that sunflower (Helianthus annuus) can produce haploid embryos spontaneously through parthenogenesis, which is when embryos form from unfertilized eggs.. Testing a chemical method used in maize to induce haploids, they observed viable seeds on emasculated, unpollinated sunflower plants. These seeds germinated into plants with smaller leaves and reduced stomatal size, confirming they were true haploids from unfertilized egg cells. Genetic analysis confirmed these seeds were maternally derived, lacking paternal DNA. By combining chemical emasculation, high-intensity light to stimulate parthenogenesis, and genome doubling, they developed a highly efficient, non-transgenic DH pipeline, reducing breeding timelines from six years to just ten months. This breakthrough not only accelerates sunflower breeding plans but also opens the door to transferring DH technology across species. Understanding the genetic and cellular mechanisms behind this process could eventually allow parthenogenesis to be induced in other crops, revolutionizing plant breeding and accelerating crop improvement. (See also the News and Views by Todesco and Reisburg 10.1038/d41586-025-00904-8) (Summary by Gourav Arora @gourav_arora_g) Nature 10.1038/s41586-025-08798-2
Hybrid crops are key to modern agriculture, offering higher yields, improved stress resilience, and greater uniformity through heterosis (“hybrid vigor”), where offspring from two different inbred lines outperform their parents. However, producing homozygous inbred lines for stable hybrid seed production traditionally takes at least six generations of self-pollination, slowing breeding progress. Doubled haploid (DH) technology offers a faster solution by generating homozygous lines in a single generation through haploid embryo formation and genome doubling. While DH systems are established in maize, their use in other crops has been limited by inefficiency and genotype dependence. In a serendipitous discovery, Lv et al. found that sunflower (Helianthus annuus) can produce haploid embryos spontaneously through parthenogenesis, which is when embryos form from unfertilized eggs.. Testing a chemical method used in maize to induce haploids, they observed viable seeds on emasculated, unpollinated sunflower plants. These seeds germinated into plants with smaller leaves and reduced stomatal size, confirming they were true haploids from unfertilized egg cells. Genetic analysis confirmed these seeds were maternally derived, lacking paternal DNA. By combining chemical emasculation, high-intensity light to stimulate parthenogenesis, and genome doubling, they developed a highly efficient, non-transgenic DH pipeline, reducing breeding timelines from six years to just ten months. This breakthrough not only accelerates sunflower breeding plans but also opens the door to transferring DH technology across species. Understanding the genetic and cellular mechanisms behind this process could eventually allow parthenogenesis to be induced in other crops, revolutionizing plant breeding and accelerating crop improvement. (See also the News and Views by Todesco and Reisburg 10.1038/d41586-025-00904-8) (Summary by Gourav Arora @gourav_arora_g) Nature 10.1038/s41586-025-08798-2
“Tea rice”- the catechin fortified rice
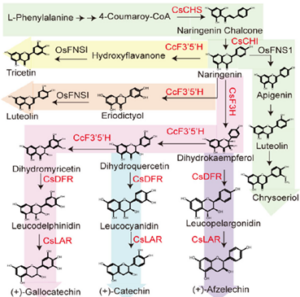 Rice feeds nearly half of the world population. Rice grains are rich in starch but low in micronutrients and bioactive compounds such as flavonoids. By contrast, tea leaves are rich in antioxidants such as catechins but low in calories. Zhu and colleagues combine the advantages of rice and tea by producing “tea rice”. They transformed six catechin synthesis genes from tea plant (Camellia sinensis) and montbretia (Crocosmia × crocosmiiflora) into rice plants and drove their expressions in the endosperm. The transgenic rice grains had increased catechins as well as other flavonoids including kaempferol and quercetin. Consistently, the transgenic rice grains also had increased antioxidating and radical-scavenging capacities. The antioxidating environment subsequently favours the accumulation of B-vitamins in the rice grains. The study sets a successful example of genetic engineering for metabolic flux redirection and multiple health-beneficial compound accumulation in rice. (Summary by Yee-Shan Ku @YeeShanKu1) Plant Biotechnol. J. 10.1111/pbi.70060
Rice feeds nearly half of the world population. Rice grains are rich in starch but low in micronutrients and bioactive compounds such as flavonoids. By contrast, tea leaves are rich in antioxidants such as catechins but low in calories. Zhu and colleagues combine the advantages of rice and tea by producing “tea rice”. They transformed six catechin synthesis genes from tea plant (Camellia sinensis) and montbretia (Crocosmia × crocosmiiflora) into rice plants and drove their expressions in the endosperm. The transgenic rice grains had increased catechins as well as other flavonoids including kaempferol and quercetin. Consistently, the transgenic rice grains also had increased antioxidating and radical-scavenging capacities. The antioxidating environment subsequently favours the accumulation of B-vitamins in the rice grains. The study sets a successful example of genetic engineering for metabolic flux redirection and multiple health-beneficial compound accumulation in rice. (Summary by Yee-Shan Ku @YeeShanKu1) Plant Biotechnol. J. 10.1111/pbi.70060
The hidden architecture of symbiosis: Casparian strips in root and nodule integration
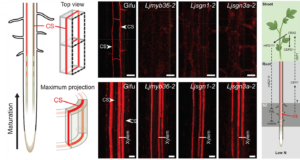 The Casparian strip (CS), a lignin-rich apoplastic barrier in the root endodermis, forms early in development and regulates solute flow between the soil and the vasculature. It allows plants to control ion uptake, defend against pathogens, and coordinate developmental signals. While its functions in these processes are well studied, its role in root nodule symbiosis has remained elusive. Recent findings by Defeng et al. reveal a strong link between CS formation and nodule development in Lotus japonicus. Mutants lacking key CS regulators (LjMYB36, LjSGN1, and LjSGN3) showed impaired lignin deposition, delayed barrier formation, and reduced nodule formation after inoculation with Mesorhizobium loti R7A. Notably, wild-type nodules contain an internal Casparian strip within their vascular endodermis, serving as a secondary barrier that regulates nutrient and signal exchange between the plant and symbiont. In CS-deficient nodules, this internal barrier was absent, leading to ion imbalances (notably sodium, potassium, molybdenum, and phosphorus). These mutants also failed to produce LjCEP1, a shoot-directed peptide induced under nitrogen starvation. Without LjCEP1, the shoot could not suppress LjTML (TOO MUCH LOVE), a negative regulator of nodulation, further impairing nodule formation. Together, these results redefine the Casparian strip as a dynamic regulator of symbiotic coordination and metabolic exchange. These findings have broad implications for enhancing nitrogen fixation in crops, potentially improving agricultural productivity and sustainability. (Summary by Gourav Arora @gourav_arora_g) Science 10.1126/science.ado8680
The Casparian strip (CS), a lignin-rich apoplastic barrier in the root endodermis, forms early in development and regulates solute flow between the soil and the vasculature. It allows plants to control ion uptake, defend against pathogens, and coordinate developmental signals. While its functions in these processes are well studied, its role in root nodule symbiosis has remained elusive. Recent findings by Defeng et al. reveal a strong link between CS formation and nodule development in Lotus japonicus. Mutants lacking key CS regulators (LjMYB36, LjSGN1, and LjSGN3) showed impaired lignin deposition, delayed barrier formation, and reduced nodule formation after inoculation with Mesorhizobium loti R7A. Notably, wild-type nodules contain an internal Casparian strip within their vascular endodermis, serving as a secondary barrier that regulates nutrient and signal exchange between the plant and symbiont. In CS-deficient nodules, this internal barrier was absent, leading to ion imbalances (notably sodium, potassium, molybdenum, and phosphorus). These mutants also failed to produce LjCEP1, a shoot-directed peptide induced under nitrogen starvation. Without LjCEP1, the shoot could not suppress LjTML (TOO MUCH LOVE), a negative regulator of nodulation, further impairing nodule formation. Together, these results redefine the Casparian strip as a dynamic regulator of symbiotic coordination and metabolic exchange. These findings have broad implications for enhancing nitrogen fixation in crops, potentially improving agricultural productivity and sustainability. (Summary by Gourav Arora @gourav_arora_g) Science 10.1126/science.ado8680
The elephant in the genome: Cryptic infection by giant viruses
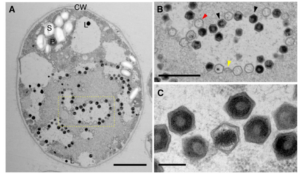 Eukaryotic genomes contain many endogenous viral elements (EVEs), which are genetic footprints of past viral infections. With the discovery of Nucleocytoplasmic Large DNA Viruses (NCLDVs), or giant viruses, research has turned to exploring how giant endogenous viral elements (GEVEs) shape host genomes. Giant viruses are prevalent in unicellular green and brown algae, and some species of bryophytes and lycophytes. While viral latency is a well-known strategy in many viral lineages, how giant viruses use latency remains poorly understood. Erazo et al. explored the effect of latency in Chlamydomonas reinhardtii green algae. Using long-read sequencing and qPCR, they discovered active GEVEs in the host genome producing new viruses, without host death. The accumulation of new viruses during the stationary phase of algal growth indicates a cryptic infection strategy. Similar viral elements have also been found in Chlamydomonas and other green algae species field isolates, suggesting that latent infections may be a widespread strategy for giant viruses. This study highlights the importance of investigating GEVEs to uncover a hidden layer of virus-host interactions that has largely gone unnoticed, with potential implications for understanding genome evolution and the broader role of giant viruses in eukaryotic biology.(Summary by Xavier Ozowara [email protected]). Science 10.1126/science.ads6303
Eukaryotic genomes contain many endogenous viral elements (EVEs), which are genetic footprints of past viral infections. With the discovery of Nucleocytoplasmic Large DNA Viruses (NCLDVs), or giant viruses, research has turned to exploring how giant endogenous viral elements (GEVEs) shape host genomes. Giant viruses are prevalent in unicellular green and brown algae, and some species of bryophytes and lycophytes. While viral latency is a well-known strategy in many viral lineages, how giant viruses use latency remains poorly understood. Erazo et al. explored the effect of latency in Chlamydomonas reinhardtii green algae. Using long-read sequencing and qPCR, they discovered active GEVEs in the host genome producing new viruses, without host death. The accumulation of new viruses during the stationary phase of algal growth indicates a cryptic infection strategy. Similar viral elements have also been found in Chlamydomonas and other green algae species field isolates, suggesting that latent infections may be a widespread strategy for giant viruses. This study highlights the importance of investigating GEVEs to uncover a hidden layer of virus-host interactions that has largely gone unnoticed, with potential implications for understanding genome evolution and the broader role of giant viruses in eukaryotic biology.(Summary by Xavier Ozowara [email protected]). Science 10.1126/science.ads6303
Friend or Foe? How fungi switch between helping and harming plants
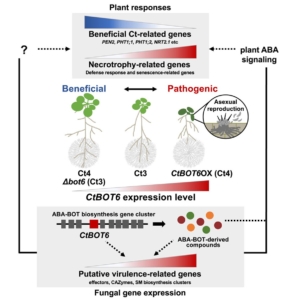 Plants coexist and interact with various microorganisms in the soil environment, including fungi. These associated fungi can not only cause diseases but also establish symbiotic interactions that boost plant health. This is the case of the endophyte Colletotrichum tofieldiae, where some strains can improve the plant’s nutritional status by transferring phosphorus from the soil, while others can exhibit pathogenic behavior. In this study, Ujimatsu and colleagues investigated the genetic causes behind this continuum between beneficial and pathogenic lifestyles. They discovered that a transcription factor called CtBOT6, part of a gene cluster involved in producing sesquiterpene metabolites that increase fungal virulence, acts like a molecular switch. Activation of this transcription factor in pathogenic isolates drives infection, while it remains inactive in beneficial ones. When CtBOT6 is overexpressed in normally non-expressing (beneficial) strains, it transforms the fungus into a necrotrophic pathogen. This shift activates virulence genes and triggers the production of a wide array of fungal compounds. The newly pathogenic fungus can now not only colonize roots but also invade and reproduce in leaves. These findings help explain how some endophytes switch between being plant allies and aggressors, providing insight into the genetic mechanisms that distinguish pathogenic and beneficial fungal lifestyles. (Summary by Carlos González Sanz @carlosgonzsanz) Curr. Biol. 10.1016/j.cub.2025.03.026
Plants coexist and interact with various microorganisms in the soil environment, including fungi. These associated fungi can not only cause diseases but also establish symbiotic interactions that boost plant health. This is the case of the endophyte Colletotrichum tofieldiae, where some strains can improve the plant’s nutritional status by transferring phosphorus from the soil, while others can exhibit pathogenic behavior. In this study, Ujimatsu and colleagues investigated the genetic causes behind this continuum between beneficial and pathogenic lifestyles. They discovered that a transcription factor called CtBOT6, part of a gene cluster involved in producing sesquiterpene metabolites that increase fungal virulence, acts like a molecular switch. Activation of this transcription factor in pathogenic isolates drives infection, while it remains inactive in beneficial ones. When CtBOT6 is overexpressed in normally non-expressing (beneficial) strains, it transforms the fungus into a necrotrophic pathogen. This shift activates virulence genes and triggers the production of a wide array of fungal compounds. The newly pathogenic fungus can now not only colonize roots but also invade and reproduce in leaves. These findings help explain how some endophytes switch between being plant allies and aggressors, providing insight into the genetic mechanisms that distinguish pathogenic and beneficial fungal lifestyles. (Summary by Carlos González Sanz @carlosgonzsanz) Curr. Biol. 10.1016/j.cub.2025.03.026
Stopping citrus greening with peptide therapy
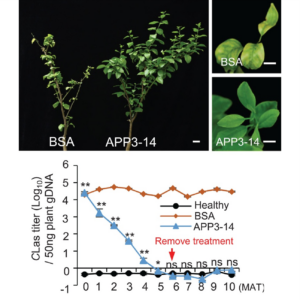 Citrus greening disease (also known as Huanglongbing) has had a huge impact on citrus fruit production worldwide, with Florida particularly hard hit. The disease is caused by insect-vector-spread bacteria, including Candidatus Liberibacter asiaticus (CLas). There is some genetic variability in susceptibility, which led Zhao et al. to investigate the role of an E3-ubiquitin ligase, PUB21, in disease susceptibility. Higher levels of PUB21, through both natural variation and induced expression, increased susceptibility. They also identified a naturally occurring dominant-negative PUB21 allele that encodes a variant without ubiquitin-ligase activity, and showed that it positively correlates with disease resistance. When they screened for potential targets of PUB21 ubiquitination, they identified the transcription factor MYB2, which regulates signaling by the defense hormone jasmonate. They also identified a bacterial effector that promotes the interaction between PUB21 and MYB2, leading to increased susceptibility. Finally, they identified an antiproteolysis peptide (APP), APP3-14, that disrupts the interaction between PUB21 and MYB2 in the presence of the bacterial effector. Remarkably, application of APP3-14 to infected trees increases MYB2 expression and jasmonate defenses, thereby decreasing bacterial growth and disease progression. This exciting work opens many avenues for control of citrus greening disease. (Summary by Mary Williams @PlantTeaching.bsky.social) Science 10.1126/science.adq7203
Citrus greening disease (also known as Huanglongbing) has had a huge impact on citrus fruit production worldwide, with Florida particularly hard hit. The disease is caused by insect-vector-spread bacteria, including Candidatus Liberibacter asiaticus (CLas). There is some genetic variability in susceptibility, which led Zhao et al. to investigate the role of an E3-ubiquitin ligase, PUB21, in disease susceptibility. Higher levels of PUB21, through both natural variation and induced expression, increased susceptibility. They also identified a naturally occurring dominant-negative PUB21 allele that encodes a variant without ubiquitin-ligase activity, and showed that it positively correlates with disease resistance. When they screened for potential targets of PUB21 ubiquitination, they identified the transcription factor MYB2, which regulates signaling by the defense hormone jasmonate. They also identified a bacterial effector that promotes the interaction between PUB21 and MYB2, leading to increased susceptibility. Finally, they identified an antiproteolysis peptide (APP), APP3-14, that disrupts the interaction between PUB21 and MYB2 in the presence of the bacterial effector. Remarkably, application of APP3-14 to infected trees increases MYB2 expression and jasmonate defenses, thereby decreasing bacterial growth and disease progression. This exciting work opens many avenues for control of citrus greening disease. (Summary by Mary Williams @PlantTeaching.bsky.social) Science 10.1126/science.adq7203
How plant vampires strike: Mechanosensitive channels in haustorium formation
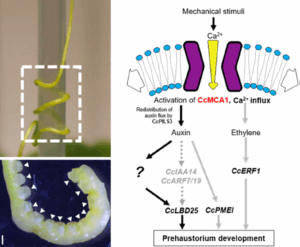
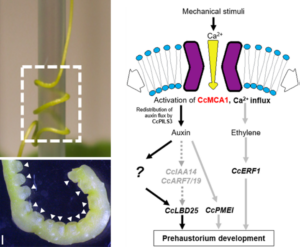 While the existence of vampires is up for debate, their botanical counterparts are very real – and much easier to find. Parasitic plants, such as Cuscuta campestris, wrap around their hosts, draining them of water and nutrients, and causing serious economic losses in agriculture. The first critical step in their attack is the formation of a haustorium – a specialized organ that attaches to the host and facilitates resource theft. Although light and mechanical cues are known to trigger haustorium primordium development, the underlying molecular mechanisms remain unclear. To address this, Park and colleagues focused on mechanosensitive ion channels (MSCs) and identified CcMCA1 as a key player in prehaustorium development. Silencing CcMCA1 significantly reduced haustorium formation, mimicking the effects of chemical inhibitors. This was accompanied by a downregulation of genes associated with haustorium development, some of which are also involved in auxin and ethylene signaling. However, the precise roles of phytohormones in this process remain to be elucidated. This study sheds light on the molecular machinery behind parasitic plant development and reveals promising targets for eliminating these “plant vampires”! (Summary by Ching Chan @ntnuchanlab) Plant Cell Physiol. 10.1093/pcp/pcaf009
While the existence of vampires is up for debate, their botanical counterparts are very real – and much easier to find. Parasitic plants, such as Cuscuta campestris, wrap around their hosts, draining them of water and nutrients, and causing serious economic losses in agriculture. The first critical step in their attack is the formation of a haustorium – a specialized organ that attaches to the host and facilitates resource theft. Although light and mechanical cues are known to trigger haustorium primordium development, the underlying molecular mechanisms remain unclear. To address this, Park and colleagues focused on mechanosensitive ion channels (MSCs) and identified CcMCA1 as a key player in prehaustorium development. Silencing CcMCA1 significantly reduced haustorium formation, mimicking the effects of chemical inhibitors. This was accompanied by a downregulation of genes associated with haustorium development, some of which are also involved in auxin and ethylene signaling. However, the precise roles of phytohormones in this process remain to be elucidated. This study sheds light on the molecular machinery behind parasitic plant development and reveals promising targets for eliminating these “plant vampires”! (Summary by Ching Chan @ntnuchanlab) Plant Cell Physiol. 10.1093/pcp/pcaf009
 Citrus greening disease (also known as Huanglongbing) has had a huge impact on citrus fruit production worldwide, with Florida particularly hard hit. The disease is caused by insect-vector-spread bacteria, including Candidatus Liberibacter asiaticus (CLas). There is some genetic variability in susceptibility, which led Zhao et al. to investigate the role of an E3-ubiquitin ligase, PUB21, in disease susceptibility. Higher levels of PUB21, through both natural variation and induced expression, increased susceptibility. They also identified a naturally occurring dominant-negative PUB21 allele that encodes a variant without ubiquitin-ligase activity, and showed that it positively correlates with disease resistance. When they screened for potential targets of PUB21 ubiquitination, they identified the transcription factor MYB2, which regulates signaling by the defense hormone jasmonate. They also identified a bacterial effector that promotes the interaction between PUB21 and MYB2, leading to increased susceptibility. Finally, they identified an antiproteolysis peptide (APP), APP3-14, that disrupts the interaction between PUB21 and MYB2 in the presence of the bacterial effector. Remarkably, application of APP3-14 to infected trees increases MYB2 expression and jasmonate defenses, thereby decreasing bacterial growth and disease progression. This exciting work opens many avenues for control of citrus greening disease. (Summary by Mary Williams @PlantTeaching.bsky.social) Science 10.1126/science.adq7203
Citrus greening disease (also known as Huanglongbing) has had a huge impact on citrus fruit production worldwide, with Florida particularly hard hit. The disease is caused by insect-vector-spread bacteria, including Candidatus Liberibacter asiaticus (CLas). There is some genetic variability in susceptibility, which led Zhao et al. to investigate the role of an E3-ubiquitin ligase, PUB21, in disease susceptibility. Higher levels of PUB21, through both natural variation and induced expression, increased susceptibility. They also identified a naturally occurring dominant-negative PUB21 allele that encodes a variant without ubiquitin-ligase activity, and showed that it positively correlates with disease resistance. When they screened for potential targets of PUB21 ubiquitination, they identified the transcription factor MYB2, which regulates signaling by the defense hormone jasmonate. They also identified a bacterial effector that promotes the interaction between PUB21 and MYB2, leading to increased susceptibility. Finally, they identified an antiproteolysis peptide (APP), APP3-14, that disrupts the interaction between PUB21 and MYB2 in the presence of the bacterial effector. Remarkably, application of APP3-14 to infected trees increases MYB2 expression and jasmonate defenses, thereby decreasing bacterial growth and disease progression. This exciting work opens many avenues for control of citrus greening disease. (Summary by Mary Williams @PlantTeaching.bsky.social) Science 10.1126/science.adq7203